Aptamers and Cancer
Download
Abstract
Oligonucleotides Aptamers are single strands of DNA and RNA with the length of 20-100 nucleotides or peptides and unique three dimensional structure that is affected by nucleotide sequence. The structure exclusively influences aptamers’ bindings with its target molecule. It reduces performance or inactivate protein and this feature is used for therapeutic purposes. In addition and through connecting to signature molecule, aptamers are used to detect specific proteins. High efficiency of aptamer technology makes them a valuable tool for diagnosing and treating different diseases including cancer. The present study is an attempt to review recent studies in this field.
Introduction
Recent reports indicate that the value of medical therapy market of nucleic acids will soon exceed US$210 billion. This figure shows the potential of these medicines for treating different diseases such as cancer, arthritis, rheumatoid, and cardiovascular disorders. A report under the title “medical therapy based on nucleic acid: the world market and its usage” categorized medical therapy based on nucleic acid into six categories and as the most important and popular medicines in the future. These six categories are antisense, intervening ribonucleic, gene therapy, nucleoside analogues, ribosomes, and aptamers. The term “aptamer” is from Latin root “aptus” for “fit.” The single strand oligonucleotides are made of RNA and DNA and usually reach the length of 20-100 nucleotide with a unique 3D structure. The structure is affected by the sequence, which also exclusively affects how aptamer is bound to its target molecules (e.g. protein with positive and negative loads, lipids, polysaccharides, antibodies, ions, viruses, tissues, cell, and small organs) at macro and micro scales [1, 2, 3, 4].
Aptamers might play a role in blocking protein-protein reactions as receptor agonist, diagnosing markers, or carrier of other materials to the cell [2]. High accuracy of aptamers enables them to detect proteins among few different amino acids [5]. After marking, aptamer probe can be used to generate a highly sensitive signal and measure the target molecule accurately[6].
In addition to efficient binding, in many cases, aptamers are featured with suppressing effects on their targets. Using them as biological diagnostic elements in biosensors is a major advance in fast and easy detection of proteins and/or other aptamer targets. The new generation of biosensors known as aptasensors are stable and perfect for clinical samples [1, 5].
There are several advantages notable about aptamers, which make them suitable for molecular diagnosis or replacing antibodies for variety of examination purposes [3]. Among these advantages, ease of design and production is notable so that an accurate and economic diagnosing system can be built easily using aptamers [7, 8].
Specificity, high tendency, and ease of binding to target molecules make aptamers capable of making exclusive binding to their target molecules ranging from pico-molar to nano-molar. This feature makes aptamer similar to monoclonal antibodies (mAbs) [2, 9]. Binding site of aptamer includes molecular grooves and gaps of the target molecules such as enzymes. Thus, aptamers demonstrate antagonistic activities and acts like many mainstream drug agents such as anti HIV virus or direct suppressors of thrombin [2].
Aptamers are not immunogen: in spite of monoclonal antibodies and peptides, aptamers do not stimulate immune system of body, which makes them more efficient in long-run [2].
Stability of aptamers in different conditions and their unlimited half-life: Aptamers demonstrate stability in a wide range of temperature and environment. Thereby, they can preserve their ability to develop the fourth unique structure of molecule. Denaturation of aptamers at high temperature is reversible, a feature that is not observed in antibodies [10, 11]. In addition and thanks to ease of changing aptamers –e.g. use of modified nucleotides, it is possible to developed resistance to nuclease in aptamers. Comparing with antibodies, another advantage of aptamers is ease of modifying them and increasing their half-life [2].
High absorbability and fast penetrability to tumors: aptamers can be used as an alternative for monoclonal antibodies in radiotherapy and imaging purposes [4].
Rapid and easy synthesis: aptamers are rapidly and easily synthesized using chemical methods, while producing monoclonal antibodies needs expensive and complicated animal and biological systems. Chemical synthesis of aptamers facilitates making variety of modifications in them. For instance, conjugating aptamers with glycol polyethylene increases half-life of aptamer [2]; or connecting two identical aptamers and dimerization increases their tendency to the target. The latter is done for anti-VEGF165 aptamer (vascular endothelial growth factor), which is a homodimer protein, and aptamer that are bound to thrombin. Thrombin binding dimmer aptamer is featured with higher suppressive activity comparing with its non-dimmer type. The aptamer suppresses thrombin and fibrinogen reactions through making a binding to thrombin so that it can demonstrate anticoagulation effects [7]. DNA aptamers in vitro are chosen and proliferated using different combinatorial chemical methods such as systematic evolution of ligands by exponential enrichment (SELEX) techniques using large libraries containing DNA synthesized random sequences (Fig.1). The libraries usually contain 1015 different sequences of oligonucleotide. However, number of the sequences depends on the length of the region. Theoretically speaking, with a variable region of 25 oligonucleotide, 425 different sequences (about 1015) in the library are assumable.
Figure 1 :SELEX technique [ 11 ]
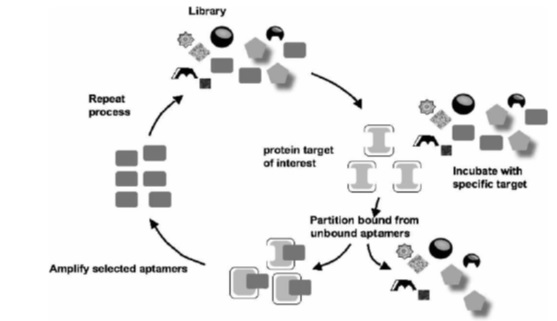
SELEX technique is a combinatorial chemistry method that was first introduced in 1990 and a great deal of development has been made in this field since then. At first, a single-stranded DNA library is mixed with a target protein; then, the DNA attached to the target protein is separated from the non-bound protein. It is then proliferated to generate a DNA library rich of a target anti-protein aptamer. This cycle is repeated for several times and by each turn, combined affinity to DNA in the library increases comparing with the last cycle. The process stops when the mixing tendency of the library stops growing, and the obtained library with highest mixing affinity is extracted as an aptamer resource. None of the problems in selection method or in-vivo generation are observed in this technique. Unlike antibody generation methods, structure and function of aptamer are preserved in this technique. In addition, SELEX technique is highly modifiable to achieve aptamers with desired specification, a feature that is not found in classic methods for generating antibodies. In addition, number and diversity of the variety in library and/or primary reservoir is higher comparing with antibodies [1, 3, 12, 13].
| Aptamer | Antibody | |
| Generation time | Less than eight weeks more or less using automatic method | More than six months |
| Specificity and combination affinity | Very high | High |
| Target controlling capability | High | Low 1/200 |
| Molecular weight | 5-8kD | 150kD |
| Immunogenicity and toxicity | None observed | Not possible |
| Target molecule situation | Intercellular and extracellular proteins | Only extracellular proteins |
| Chemical structure variability | Positive | Negative |
| Physical/chemical stability | Stable | Different |
Small size: Comparing with antibodies, aptamers are small in size and this increases surface density of the receptor and enables easier binding to analytes. [14]. Moreover, aptamers are featured with advantages comparing with RNA interference (RNAi), which are listed in Tabl 2.
| Features | Aptamer | RNAi |
| Protein | mRNA | |
| + | +++ | |
| ++ | +++ | |
| +++ | ++ | |
| Type of knockdown | +++ | - |
| Turn over/throughput | +++ | - |
| Specificity | +++ | - |
| Drug-like |
Role of aptamer is diagnosing cancers
Specificity and high efficiency of aptamers have made them a useful option in dealing with cancer in terms of diagnosis and treatment. They can be used as molecule probes to diagnose new biomarkers of cancers. There are aptamers with anti-neoplasia activity against targets inside and outside cells and at cell membrane. The main area of using these aptamers is for extracellular and intercellular targets, which is thanks to relative ease of detecting the target and penetration [2, 15].
Shangguan D et al. used a group of aptamers for specific detection of leukemia cells. The selected aptamers at nano- and picometer scale were able to specifically detect and bind with cancer cells mixed with normal cells. They used Cell-SELEX technique where aptamer was generated for the complete cells with different surface markers; then, the aptamers were specifically bound to cancer cells, selected, a proliferated. Afterward, specificity in diagnosing cancer cells was measured through marking the binding of the aptamers to the cell using florescent dye and flow cytometery. Unbound single-strand DNA source was also marked using florescent and used as negative control. Selection stage was simple and fast using small pieces of information [16].
Ferreira C.S.M et al. generated anti-protein recombinant MUC1 and measured binding capability of the aptamer to the protein using ELISA and surface plasmon resonance (SPR) methods. Using ELISA Sandwich method, the aptamer was utilized to diagnose and determine MUC1 level in buffer solutions. The surveys showed efficiency of the generated aptamer as receivers for MUC1 with very small amount of the protein [15].
Chen HW et al. used an aptamer based method to diagnose molecular markers of specific small-cell lung cancer (SCLC) with very bad phenotypes. Aptamers of the SCLC cells were generated using Cell-SELEX technique. When used for different lung cancer cells, the aptamers were bound to SCLC cells with high affinity and specificity and when coagulated with florescent nano-particles, the compounds were effective in diagnosing and differentiating SCLC cells among other cells. The survey indicated importance and potential of aptamer technique for early diagnosis of lung cancer [17].
Aptamer selection and generation technique based on cells is highly promising in development of specific molecule probes for diagnosing and selection of new cancerous biomarkers and affected cells [16].
Role of aptamers in diagnosing cancers
Major advances in treating cancer using aptamers have been in nucleoline anti-protein aptamers. The protein weighs 76KD with variety of functions in the cells including rRNA transcription and processing, simulating and recombining DNA, and stabilizing mRNA. In addition, the protein is probably effective on angiogenesis of tumor and suppressing apoptosis [18, 19, 20].
Apparently, anti-nucleoline aptamer known as As-1411 has anti-neoplasmic effects. Suppressing effects against several lines of cancer have been demonstrated in vitro. There are speculation that As-1411 demonstrates anti-tumor activity in vitro and in xenograft model of nude murine tumor [20].
Another objective sought after in using aptamer is treatment of PDGF and β-type in particular. This factor plays a role in angiogenesis. Sennino B et al. examined the role of PDGF-β in growth of tumor in mice lung carcinoma and observed that suppressing PDGF- β led to decrease in concentration of veins. Their results showed that aptamer Ax-102 functions against PDGF- β and attenuates veins in the tumor up to 79% [21].
Probably, suppressing VEGF, angiotensin 2 (ANG2), and PDGF using aptamers as independent factors or combined treatment would be a part of anti-angiogenesis strategy in the future.
NFkB is a nucleus transcription factor that plays a role in regulating several genes effective on apoptosis. The factor is a key mediator in cells inflammatory reaction that functions through generating cytokine. It appears that pre-inflammation stage, triggered by NFkB, plays a key role in progress of cancer [22, 23].
Mi J et al. used an RNA aptamer known as Ad-A-P50 that targets NFkB, which had been sensitized to apoptosis pathway depended on αTNF in vitro by adenocarcinoma of human lung [24]. In another study, they used the same aptamer to examine anti-tumor effect of Doxorubicin. Suppression of NFkB by the aptamer was coincident with increase in tumor response to the drug. This finding indicated possibilities of using the aptamers as a co-factor to improve cells’ response to chemotherapy [25].
VEGF is an epidermal growth factor with pre-angiogenesis effects such as mitogen activities on endothelial cells [26].
A notable advance in cancer treatment is generation and examination of an anti-VEGF RNA aptamer known as Pegaptanib. The aptamer is not a factor for treating cancer, however, examining its expression might lead to development of the first commercial aptamer and preparation of the ground for development of other aptamer for medical purposes. Anti-VEGF aptamers such as bevacizumab (Avastin) is a monoclonal antibody, which can be used as an anti-cancer medicine at clinical level [27].
Osteopontin is a glycoprotein factor that improves tumor cell invasion and its expression increases in several types of cancerous cells such as chest carcinoma and colorectal. Secretion of this factor increases during metastasis [27, 28].
Mi Z et al. generated an RNA aptamer of OPNR-3 type that suppressed signaling pathway of Osteopontin by entering chest cancerous cells. In addition, the aptamer caused notable decrease in tumor development and metastases in a xerograph model of chest cancer [29].
RAS is an effective oncogene in several types of cancer with notable role in apoptosis and controlling cell cycle. Tanaka Y et al. produced an anti-K-RAS RNA aptamer that prevented attachment of K-RAS to cellular membrane in vitro; K-RAS is needed for transfer of K-RAS signal. At any rate, there is a need for further in vivo and in vitro studies for thorough examination of suppressive effects of the aptamer on K-RAS [30].
β-Catenin is a multi-purpose protein that plays a role in tumorigeneses and transcription of oncogenes such as Cyclin D1 and C-myc and regulating frequent processing of oncogene mRNA transcripts [31, 32].
LeeKh et al. used an anti- β-catenin RNA aptamer in colon cancer cells. The aptamer suppressed transcript of β-catenin in Cyclin D1 and C-myc and interrupted β-catenin sequenced process in cancer cells. In addition, it suppressed cell cycle and reduced cells’ tumor formation potential in vitro [32].
HER2 and HER3 are homologues of human epidermal growth factor receptor (EGF) that function as surface receptor with kinase activity and play a role in proliferation and differentiation. Over-expression of kinases in several solid tumors have been observed in breast and ovarian cancer. Suppressing them is considered as a key clinical intervention, as they are observed in Herceptin (HER2). Researchers in California University developed RNA aptamers that can bind to the 2nd extracellular of HER2 and HER3. They determined a specific aptamer that can control HER2 and HER3 in breast cancer cells. The aptamer is a clinical element for developing cancer drugs [9].
Receptor tyrosine kinases (RTKs), in large number, play a role in signaling stage of cells growth and proliferation regulation of several cancers. Thereby, it is studied widely by cancer researchers. In this field, rearranged during transfection (RET) are notable. Germline mutation in RET gene induces permanent activity in the receptor and creates 2A and 2B multiple endocrine neoplasia (MEN) and thyroid carcinoma [33].
Cerchia L et al. developed an RET aptamer at cancerous cell surface, which not only binds to and diagnose RET, but also suppresses downstream RET [33].
Capability of aptamers for accurate targeting and ease of modifying and adjusting them have resulted in several new therapeutic and diagnostic methods based on the aptamers. In this regard, development of conjugates of nanoparticles-aptamer and/or using aptamer as carrier of drugs to cells (e.g. SiRNA chimera aptamer) are notable [34, 35, 36].
Huang YF et al. examined the effect of PDGF anti-marker of gold nanoparticle-aptamer conjugates on breast cancer cells. The cancer cells with more markers comparing with normal cell had more interactions with this conjugates and more gold particles entered these cells. The agents suppressed proliferation of cancer cells; however, they were ineffective on normal cells [34].
Dhar S et al. used an RNA aptamer of Prostate specific membrane antigen (PSMA) with a polylacticglycolic acid polymer EPG to add Cisplatin to prostate cancerous cells. Comparing with cisplatin used independently, the structure showed higher suppressing effects on cancer cells [35].
Wullner U et al. is a SiRNA chimera – non-valent aptamer- containing two anti-PSMA aptamers and one siRNA that suppresses eukaryotic elongation factor 2 (EEF2) and induces apoptosis. This indicates that this factor can be considered as a target in cancer treatment. Chimera structure expresses PSMA antigen on cancerous cells and demonstrates toxic effects [36].
It is notable that macugene is the first FDA approved and successful aptamer-based drug, which actually is an anti-vascular endothelial growth factor (VEGF 165) for treatment of age related macular degeneration (AMD). Other aptamer drugs are at clinical or laboratory research phases.
The reviewed studies indicated high potential and importance of aptamers in diagnosing and treating cancers. It is notable, however, that developing practical diagnosis or treatment methods using aptamers entails with further surveys and studies.
References
[1]. Strehlitz B, Nikolaus N, Stoltenburg R. Protein Detection with Aptamer Biosensors. Sensors (Basel). 2008;8(7):4296-307.
[2]. Javaherian S, Musheev MU, Kanoatov M, Berezovski MV, Krylov SN. Selection of aptamers for a protein target in cell lysate and their application to protein purification. Nucleic acids research. 2009;37(8):e62.
[3]. Barbas AS, White RR. The development and testing of aptamers for cancer. Curr Opin Investig Drugs. 2009;10(6):572-8.
[4]. Pieve C Da IJ, Parkins A, Missailidis S. Development of anti-Muc1 DNA aptamers for the imaging and radiotherapy of breast cancer. Breast Cancer Research .2006;8:(suppl 2).
[5]. Collett JR, Cho EJ, Ellington AD. Production and processing of aptamer microarrays. Methods. 2005;37(1):4-15.
[6]. Yilin L LG, Zhao Yang Z, Jijun T, Jianwei X. Recent advances of aptamer sensors. Science in China Series B; Chemistry March. 2008;51(3):193-204.
[7]. Hasegawa H, Taira KI, Sode K, Ikebukuro K. Improvement of Aptamer Affinity by Dimerization. Sensors (Basel). 2008;8(2):1090-8.
[8]. Wu ZS, Zheng F, Shen GL, Yu RQ. A hairpin aptamer-based electrochemical biosensing platform for the sensitive detection of proteins. Biomaterials. 2009;30(15):2950-5.
[9]. HER2 and HER3 Aptamers [database on the intrnet].The Regents of the University of California (UCLA): 2009: Available from: http:// www.ibridgenetwork.org/ucla/her2-and-her3-aptamers.
[10]. Pestourie C, Tavitian B, Duconge F. Aptamers against extracellular targets for in vivo applications. Biochimie. 2005;87(9-10):921-30.
[11]. Lee JO, So HM, Jeon EK, Chang H, Won K, Kim YH. Aptamers as molecular recognition elements for electrical nanobiosensors. Analytical and bioanalytical chemistry. 2008;390(4):1023-32.
[12]. Tombelli S, Minunni M, Mascini M. Analytical applications of aptamers. Biosens Bioelectron. 2005;20(12):2424-34.
[13]. Nitsche A, Kurth A, Dunkhorst A, Panke O, Sielaff H, Junge W, et al. One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotechnol. 2007;7(1):48.
[14]. Balamurugan S, Obubuafo A, Soper SA, Spivak DA. Surface immobilization methods for aptamer diagnostic applications. Anal Bioanal Chem. 2008;390(4):1009-21.
[15]. Ferreira CS, Papamichael K, Guilbault G, Schwarzacher T, Gariepy J, Missailidis S. DNA aptamers against the MUC1 tumour marker: design of aptamer-antibody sandwich ELISA for the early diagnosis of epithelial tumours. Anal Bioanal Chem. 2008;390(4):1039-50.
[16]. Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(32):11838-43.
[17]. Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L, et al. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem. 2008;3(6):991-1001.
[18]. Srivastava M, Pollard HB. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13(14):1911-22.
[19]. Hovanessian AG, Puvion-Dutilleul F, Nisole S, Svab J, Perret E, Deng JS, et al. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Experimental cell research. 2000;261(2):312-28.
[20]. Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther.2006;5(12):2957-62.
[21]. Sennino B FB, Mc Cauley D, Le T, Mc Cauley T, al KJe. Sequential loss of tumour vessel pericytes and endothelial cells after inhibition of platelet-derived growth factor B by selective aptamer AX102. Cancer Res. 2007;67(15):7358-67.
[22]. Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nature reviews Cancer. 2005;5(4):297-309.
[23].Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nature immunology. 2002;3(3):221-7.
[24]. Mi J, Zhang X, Liu Y, Reddy SK, Rabbani ZN, Sullenger BA, et al. NF-kappaB inhibition by an adenovirus expressed aptamer sensitizes TNFalpha-induced apoptosis. Biochem Biophys Res Commun. 2007;359(3):475-80.
[25]. Mi J ZX, Rabbani ZN, Liu Y, Reddy SK, SU Z, et, al. KJe. RNA aptamer-targeted inhibition of NF-kB supresses non-small cell lung cancer resistance to doxorubicin. Mol Ther. 2008;16(1):66-73.
[26]. Kim KJ, Li B, Winer J, Armanini M, Gillett N, Phillips HS, et al. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362(6423):841-4.
[27]. Ng EW, Shima DT, Calias P, Cunningham ET, Jr., Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5(2):123-32.
[28]. Weber GF, Ashkar S, Cantor H. Interaction between CD44 and osteopontin as a potential basis for metastasis formation. Proceedings of the Association of American Physicians. 1997;109(1):1-9.
[29]. Mi Z, Guo H, Russell MB, Liu Y, Sullenger BA, Kuo PC. RNA aptamer blockade of osteopontin inhibits growth and metastasis of MDA-MB231 breast cancer cells. Mol Ther. 2009;17(1):153-61.
[30]. Tanaka Y, Akagi K, Nakamura Y, Kozu T. RNA aptamers targeting the carboxyl terminus of KRAS oncoprotein generated by an improved SELEX with isothermal RNA amplification. Oligonucleotides. 2007;17(1):12-21.
[31]. Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398(6726):422-6.
[32]. Lee HK, Choi YS, Park YA, Jeong S. Modulation of oncogenic transcription and alternative splicing by beta-catenin and an RNA aptamer in colon cancer cells. Cancer Res. 2006;66(21):10560-6.
[33]. Cerchia L, Duconge F, Pestourie C, Boulay J, Aissouni Y, Gombert K, et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 2005;3(4):e123.
[34]. Huang YF LY, Lin ZH, Chang HT. Aptamer modified gold nanoparticles for targeting breast cancer cells through light scattering. J Nanopart Res. 2009;11(4):775-83.
[35]. Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc Natl Acad Sci U S A. 2008;105(45):17356-61.
[36]. Wullner U NI, Eller A, Kleines M, Tur MK, Barth, S. Cell-specific induction of apoptosis by rationally designed bivalant aptamer-si RNA transcripts silencing eukariotic elongation factor 2. Curr Cancer Drug Targets 2506. 2008;8(7):554-65.
License
Copyright
© ,
Author Details