Disruption of FGF2-Heparin-FGFR1 Complex Formation by Antiangiogenic Small Molecule Found In Silico
Download
Abstract
Background: Basic fibroblast growth factor (FGF2) is one of the most potent proangiogenic proteins involving in tumor angiogenesis. Interaction of heparin with FGF2 and FGF2 receptor (FGFR1) form a ternary complex that is prerequisite for FGF2 proangiogenic activity. Therefore, this interaction can be an important target for inhibition of angiogenesis.
Material and Method: In this study, we performed screening studies by computer-aided techniques to find a small molecule interfering with this interaction. Based on ionic interactions, we found seventeen small molecules which had the capability of angiogenic inhibition. According to important negative charge distances, benzene-1, 2, 4-tricarboxylic acids, known as Trimellitic acid (TMLA), was chosen. MTT viability test, real time PCR, tube formation assay, and Flowcytometry technique were used to evaluate TMLA effect.
Results: Here, the viability of HUVECs was decreased following exposure to TMLA only at high concentrations. According to real time PCR, gene expression was dramatically decreased in comparison to negative control, indicating that TMLA is an effective agent on reduction of CD31 expression. At half maximal inhibitory concentration (13mM), TMLA inhibited HUVEC tube formation process. Annexin V-FITC / PI flow cytometry technique revealed that TMLA inhibitory effect was via apoptosis.
Conclusion: With respect to findings of the present study, it is possible to extend FGF2 inhibitors with much specificity based on ionic interactions strategy.
Materials and Methods
In this study, we performed screening studies by computer-aided techniques to find a small molecule interfering with this interaction. Based on ionic interactions, we found seventeen small molecules which had the capability of angiogenic inhibition. According to important negative charge distances, benzene-1, 2, 4-tricarboxylic acids, known as Trimellitic acid (TMLA), was chosen. MTT viability test, real time PCR, tube formation assay, and Flowcytometry technique were used to evaluate TMLA effect.
Results
Here, the viability of HUVECs was decreased following exposure to TMLA only at high concentrations. According to real time PCR, gene expression was dramatically decreased in comparison to negative control, indicating that TMLA is an effective agent on reduction of CD31 expression. At half maximal inhibitory concentration (13mM), TMLA inhibited HUVEC tube formation process. Annexin V-FITC / PI flow cytometry technique revealed that TMLA inhibitory effect was via apoptosis.
Conclusion
With respect to findings of the present study, it is possible to extend FGF2 inhibitors with much specificity based on ionic interactions strategy.
Introduction
Angiogenesis is a physiological process involving in the sprouting of nascent vasculature from existing blood vessels. It is now widely recognized that angiogenic activity is crucial in the progress of tumors and metastasis [1, 2]. Therefore, angiogenesis inhibition has become an appealing therapeutic strategy for cancer management [3]. The tumors recruit blood vessels from the surrounding tissue to induce the growth of blood vessels to supply them with oxygen and nutrients. Basic fibroblast growth factor (FGF2) is one of the most potent proangiogenic proteins which has been shown to influence proliferation, migration, and differentiation of cells [4-7]. This growth factor needs to interact with heparin or heparan sulfate proteoglycans (HSPGs) molecules for binding to cell surface tyrosine kinase receptor. This interaction is a prerequisite for this growth factor to recognize its’ specific site to the signaling receptor (FGFR1) to form a trimolecular complex of FGF2-HSPG-FGFR1 and receptor dimerization [4, 8]. Therefore, this interaction can be an important target for the inhibition of angiogenesis [2, 9, 10]. HSPGs are heparin like molecules widely distributed on cell surfaces and extracellular matrix [11-13]. It is composed of alternating units of sulfated glucuronic acid and glucosamine derivatives. Besides anticoagulant activity, it interacts with the proangiogenic proteins in tumor angiogenesis [14, 15]. Not surprisingly, HSPGs and their fragments modulate the onset and development of cancer [16]. It has been suggested that in some growth factors like FGF2, besides HSPG, binding of adenosine three phosphate (ATP) is also essential for their biological activity [17].
Amino acids, which are located between residues 128–144 of FGF2, forming the part of heparin binding domain (HBD) that is identified as the crucial physiologically active part of this molecule [18]. K128, R129, K134, K138, and K144 positively charge amino acids of this dominant part, and can bind to phosphate groups [18, 19]. Rose et al., (2010) performed site directed mutagenesis of positively charged amino acids in the HBD of FGF2 (aa 128–144) to find out whether it reduces ATP binding affinity or not. They observed a dramatic reduction of the autoradiographic signal intensity with the triple mutant FGF2 (K128A/R129A/K134A). Moreover, ATP binding to FGF2 protects it from physical and thermal degradation and conserves it from proteolytic cleavage.
Additionally, the conformational changes were observed after ATP binding to FGF2.
The aim of the present research was to find a small molecule with antiangiogenic effect through interfering with these interactions using computer-aided techniques.
Materials and Methods
Bioinformatics and Screening Studies
A heparin that binds to both FGF2 and FGFR1 will act as a stimulator and a heparin that only binds to FGF2 will act as an inhibitor of signaling by sequestering the growth factor [21, 22]. Therefore, the difference between heparin interaction in FGF2-Heparin and FGF2-Heparin-FGFR1 complexes is crucial to be considered. Thus, we analyzed the co-crystal structures of the FGF2–Heparin and FGF2–Heparin- FGFR1 to find this difference. The crystal structure of FGF2, FGF2–FGFR1, and ternary complex of FGF2-Heparin-FGFR1 were retrieved from the RCSB Protein Data Bank and chosen as the structure of the reference protein. The distances between positively charged amino acids of the target regions of FGF2 and the receptor, where they interact with heparin, were measured by Swiss-PdbViewer 4.0.3.
FGF2 and FGFR1 Amino Acids Which Interact with Heparin
Amino acids of FGF2 which interact with heparin included: N27, R120, T121, K125, K129, Q134, K135, and A136 (PDB ID: 1BFC) (http://www.pdb.org/pdb/home/ home.do). The measured distances between positively charged amino acids of FGF2 are shown in Tabl.1.
| Amino acids | FGF2 with H no FGFR1 | FGF2 no H (A) with FGFR1 | FGF2 with H (A) with FGFR1 | FGF2 no H (B) with FGFR1 | FGF2 with H (B) with FGFR1 |
| N27-R120 | 6.72 | 5.29 | 5.29 | 5.1 | 5.82 |
| N27-T121 | 7.64 | 7.74 | 7.69 | 7.95 | 6.9 |
| N27-K125 | 5.83 | 6.46 | 5.69 | 6.4 | 5.88 |
| N27-K129 | 12.27 | 10.5 | 12.9 | 9.46 | 9.72 |
| N27-Q134 | 7.14 | 9.37 | 7.12 | 9.5 | 7.46 |
| N27-K135 | 7.07 | 6.51 | 8.48 | 6.66 | 5.94 |
| N27-A136 | 3.96 | 4.08 | 4.05 | 3.86 | 4.69 |
| R120-T121 | 8.86 | 9.19 | 6.92 | 9.39 | 6.64 |
| R120-K125 | 11.16 | 9.22 | 7.41 | 9.05 | 7.3 |
| R120-K129 | 15.82 | 12.65 | 11.89 | 11.51 | 8.54 |
| R120-Q134 | 12.86 | 12.15 | 9.65 | 12.02 | 8.95 |
| R120-K135 | 7.64 | 5.09 | 9.73 | 5.00 | 8.48 |
| R120-A136 | 10.32 | 8.8 | 8.87 | 8.34 | 9.53 |
| T121-K125 | 10.17 | 9.99 | 9.88 | 10.26 | 7.21 |
| T121-K129 | 11.35 | 8.87 | 11.34 | 8.93 | 8.75 |
| T121-Q134 | 12.46 | 13.74 | 12.2 | 14.4 | 11.07 |
| T121-K135 | 13.57 | 13.02 | 15.23 | 13.15 | 11.9 |
| T121-A136 | 10.82 | 10.71 | 10.58 | 10.72 | 10.29 |
| K125-K129 | 8.02 | 6.22 | 8.78 | 5.04 | 4.41 |
| K125-Q134 | 2.55 | 3.82 | 2.58 | 4.19 | 4.18 |
| K125-K135 | 11.16 | 11.14 | 11.22 | 11.46 | 11.11 |
| K125-A136 | 5.34 | 5.86 | 5.08 | 5.91 | 5.71 |
| K129-Q134 | 9.66 | 8.58 | 10.24 | 8.07 | 6.32 |
| k129-k135 | 18.54 | 16.09 | 19.29 | 15.19 | 14.6 |
| K129-A136 | 12.94 | 11.01 | 13.27 | 10.03 | 9.99 |
| Q134-K135 | 11.25 | 12.97 | 11.39 | 13.29 | 10.95 |
| Q134-A136 | 5.13 | 7.26 | 4.96 | 7.56 | 5.42 |
| K135-A136 | 7.65 | 8.32 | 9.41 | 8.37 | 7.97 |
Additionaly, there are five lysines that form the most crucial part of FGFR1 in this interaction:
K160, K163, K172, K175, and K177 (PDB ID: 1FQ9)
We also measured the distances between related positively charged amino acids in FGF2-FGFR1 complex
in which heparin was absent (PDB ID: 1CVS). (PDB ID: 1BFC) file is FGF2 monomer complex with just heparin (hexamer fragment) and not with receptor; the condition in which the signaling does not occur. While A and B are two chains of ligand, the chains of receptor are C and D in dimer interactions of (PDB ID: 1FQ9) and (PDB ID: 1CVS) files. FGF2-FGFR1 is in complex with octamer fragment of heparin in 1FQ9 file, the condition that 2(FGF2-Heparin-FGFR1) has formed. Validation of reported files was checked at https://validate.wwpdb.org/validservice/.
Sulfurs of Heparin in FGF2-Heparin and FGF2-Heparin-FGFR1 Complexes
Negative charge distances between intramolecular sulfurs of heparin were measured by Swiss-PdbViewer 4.0.3 (Tabl 2). PDB entry 1BFC (complex of FGF2-Heparin) and PDB entry 1FQ9 (FGF2-Heparin-FGFR1) were used for this aim (all data are not shown).
| Distance (Å) | ||||||||
| Heparin(6) | 13.7 | 13.5 | 11.1 | 11.1 | 10.7 | 7.6 | 7.4 | 7.3 |
| Heparin(8) | 15.2 | 10.8 | 12.6 | 12.6 | 12.9 | 10.0 | 4.6 | 4.6 |
ATP Like Drugs Which Disrupt Binding of Heparin
As HBD of FGF2 also interacts with ATP, we searched ATP like drugs which can interrupt binding of heparin to FGF2. They included fosfonet, foscarnet [24], and bisphosphonates such as etidronate and clodronate [25]. Molecular structures of these drugs were drawn by Marvin sketch and optimized by Marvin space 5.9.3, respectively. Negative charge distances were measured by Swiss-PdbViewer 4.0.3 in the final step (Tabl 3).
| Distance (Å) | |||||||||
| Foscarnet | 4.9 | 4.5 | 4.4 | 3.7 | 3.7 | 3.0 | 2.6 | 2.5 | 2.5 |
| Fosfonet | 4.9 | 4.5 | 4.4 | 3.7 | 3.7 | 3.0 | 2.6 | 2.5 | 2.5 |
| Etidronate | 4.8 | 4.7 | 4.6 | 3.9 | 3.9 | 3.9 | 3.1 | 3.1 | 3.0 |
| Clodronate | 5.4 | 4.8 | 4.7 | 4.7 | 4.7 | 4.3 | 4.2 | 3.9 | 3.0 |
a) Fosfonet, Foscarnet, Etidronate and Clodronate are ATP like drugs could interrupt binding of Heparin to FGF2 by interacting with HBD. Molecular structures of these drugs were drawn by Marvin sketch and optimized by Marvin space 5.9.3 applications respectively. Negative charge distances were measured by Swiss-PdbViewer 4.0.3.
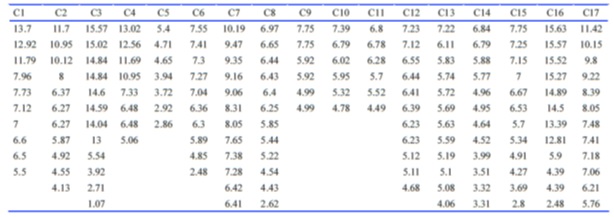
Considering that three negative charges and some special distances are common properties of these molecules, we searched biomolecules containing three negative charges in http://zinc.docking.org database. Among those molecules, some of them were excluded due to the lack of stability, the solubility problem, or complexity of structure. Seventeen molecules were reminded that were drawn by Marvin sketch. The 2D structure of all given molecules were conformed to 3D structure by Marvin space 5.9.3 using molecular mechanics method based on the dreiding force field to make approximate starting structures. Then, distances between negative charges were measured by Swiss-PdbViewer 4.0.3 (Table 4) or (Fig.6).
Figure 6 (Table 4): A: Popular Names and Structures of Three Negative Charge Compounds Found at Zinc Data Base (A) and Intramolecular Negative Charge Distances of Them (B).
Cell Culture
Adherent cell lines of human umbilical vein endothelial cell (HUVEC) and A172 cell line were obtained from national cell bank of Pasteur institute of Iran and grown in Dulbecco’s Modified Eagle’s Media (DMEM; Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (national cell bank of Pasteur institute of Iran) in 37° C humidified incubator at 5% CO2. Nearly confluent cells were passaged using 0.25% trypsin and medium changed every 3 days. Two cell lines (HUVEC and A172) for MTT assay and one cell line (HUVEC) for other assays were used.
MTT Viability Assay
Cell viability of two cell lines was evaluated using MTT assay. Briefly, 1.5* 104 cells/wells were seeded in 96-well plates and incubated for 24h at 37°C. 50µl of Trimellitic acid (TMLA) at 8 and 4 different concentrations was added to each well of HUVEC and A172 (with low FGFR expression) cell lines, respectively, in triplicate. After 24 h treatment, the medium was replaced with 100µl of MTT (3-(4, 5-dimethylthiazol -2- yl)- 2, 5-diphenyltetrazolium bromide) and incubated for 4 h in the dark. After incubation, MTT solution was removed, 100 µl isopropanol was added to each well, and incubated for additional 30 mins in the dark. The reactive product was measured at 570 nm with a reference absorbance in 630 nm by an ELISA reader. The assay was performed in triplicate for both cell types.
RNA Extraction and Real Time PCR
RNeasy Plus mini kit (Qiagen, USA) was used to purify total RNA according to the manufacture’s instruction; diluted with 30 µl RNAse free water, and quantified on a spectrophotometer instrument (NanoDrop, Eppendorf, Germany). One strand DNA was generated using Prime Script RT Reagent Kit (TaKaRa, Japan). Primers were designed using three different software packages, including Gene Runner v. 3.05, Primer Express v. 2.5, and Beacon Designer v. 7.5. Primer sequences of CD 31 for PCR amplification were (F: 5’-TCAAGCCTCAGCACCAGA-3’) and (R: 5’-GCACTCCTTCCACCAACAC-3’). Real time PCR (qPCR) was performed using SYBR Premix Ex Taq II master mix (TaKaRa, Japan) on one step instrument (Applied Bio systems, USA). Five concentrations of TMLA (0.1, 0.5, 12, 13, and 14 mM) and one concentration (0.5 mM) of vitamin C were considered as positive control [26], applying for gene expression on HUVECs. Housekeeping gene encoding GAPDH was used as endogenous control. Specific GAPDH primer sequences were (F: 5’-GAGTCCACTGGCGTCTTCA-3’) and (R: 5’-TCTTGAGGCTGTTGTCATACTTC-3’). The following thermal condition was carried out for amplification: 95˚ C for 15 sec as holding time, 95˚ C for 5 sec and 60˚ C for 30 sec in each cycle. Fluorescence was collected in annealing-extension time in each cycle. Melting curve analysis was carried out in three steps: 95 ˚C, 60˚ C and stepwise heated to 95˚ C with a ramp rate of 0.3˚ C.
HUVEC Tube Formation Assay
Tube formation assay was done according to manufacture instruction (Chemicon International, MA, USA). Briefly, Matrix gel was diluted with staining buffer in 1:9 ratio. 50µl of gel was added in each 96 deep well plate. In order to scaffold generation, the plate was incubated in a 37° C humidified CO2 incubator for 1 h. 104 HUVECs and diluted TMLA in special medium were added to the wells and kept in incubator for additional 12 h. Tube generation was evaluated every 2 h using inverted fluorescent microscope. After 12 h, the medium was discarded and wells were washed twice with PBS. 200 µl of staining solution, Calcein AM (BD Biosciences), was added to each well and the plate was transferred to incubator for 30 mins. PBS and VEGF were used as negative and positive controls, respectively.
Apoptosis Assay by Flow Cytometry
Huvec cells were seeded in triplicate into 6-well plates (2*104 cells /well) and incubated at 37° C to reach 70-80% confluency. Cells were then treated with TMLA at three concentrations (5, 10, and 13 mM) and incubated for 24 h. Apoptosis was analyzed by annexin V-FITC / PI double staining according to the manufacturer’s protocol (IQ products, Netherland). After harvesting, the cells were washed twice with calcium buffer 1X and re-suspended in 100 µL calcium buffer. 10µL annexin V-FITC was added to the cells and incubated at 4° C for 20 minutes in the dark. The cells then were washed with calcium buffer and 10 µl PI was added. The samples were incubated at 4° C for 10 mins in the dark and then subjected to flow cytometry (Partec, Germany) evaluation.
Statistical Analysis
Normal distribution of all data was checked by Shapiro-wilk test. Then, all data were statistically evaluated by analysis of variance (ANNOVA) followed by Tukey’s using SPSS (version 16.00). P<0.05 was considered statistically significant. The results were expressed as mean ± SEM from three independent experiments. IC50 was obtained using Auto Graph pad Instant 3.00.
Results
Bioinformatics
Negative Charge Distances of ATP Like Drugs
Comparing four antiangiogenic drugs (etidronate, clodronate, foscarnet, and fosfonet), based on ATP binding domain, indicated that distances of 4.87 and 2.49 were common distances in foscarnet and fosfonet. Common distances in clodronate and etidronate were 4.75 and 2.97. Among seventeen molecules we found in zinc database (Table 4), five of them (C5, C6, C8, C15, C16) had almost those two distances simultaneously. Benzene-1, 2, 4-tricarboxylic acids (Trimellitic acid) was chosen for availability and lack of toxicity . The = distances of 2.48 and 4.85 were considered for TMLA.
Sulfurs Distances (negative charges) of Heparin in FGF2-Heparin and FGF2-Heparin-FGFR1 Complexes
Comparing the negative charge distances of heparin in both complexes of FGF2-Heparin and FGF2-Heparin-FGFR indicated that some of distances between sulfurs of heparin in FGF2-Heparin were different from those between sulfurs of FGF2-Heparin-FGFR (Table 2). For example, distances of 7.65, 7.42, and 7.35 between sulfurs of heparin in Heparin-FGF2 were different from those between sulfurs of heparin in FGF2-Heparin -FGFR1; that were 10, 4.75, and 4.61 respectively. This obvious difference suggested that the mentioned distances probably were important in the mode of interaction, the interaction of heparin with ligand, or both ligand and receptor. The numbers of 7.55, 7.41, and 7.30 were intramolecular negative charge distances of TMLA that were approximately the same as those possibl important numbers of heparin that were in binding with just FGF2.
Negative charge distances of intramolecular sulfurs of Heparin were measured by Swiss-PdbViewer 4.0.3. Heparin (6): complex of FGF2 with Heparin hexamer, the condition in which the signaling does not occur (PDB entry 1BFC). Heparin (8): complex of FGF2 -FGFR1 with Heparin octamer, the condition that 2(FGF2-Heparin-FGFR1) has formed (PDB entry1FQ9).
FGF2 Amino Acids Which Interact with Heparin
Positive charge distances between K135 and R120 and between K135 and A136 of FGF2 in Heparin-FGF2 complex were 7.64 and 7.65, respectively; whereas, positive charge distances between those amino acids were obviously different in the presence of the receptor (Table 1) . As the residue K135 is essential for heparin-binding and it is in direct contact with sulfur anions of heparin [23], presumably sulfur anions of heparin with a distance of Fosfonet, Foscarnet, Etidronate, and Clodronate are ATP like drugs that could interrupt binding of Heparin to FGF2 by interacting with HBD. Molecular structures of these drugs were drawn by Marvin sketch and optimized by Marvin space 5.9.3 applications, respectively. Negative charge distances were measured by Swiss-PdbViewer 4.0.3.
7.65 interact with positive charges, bridging between K135 and R120 or between K135 and A136.
Cytotoxicity assay (MTT)
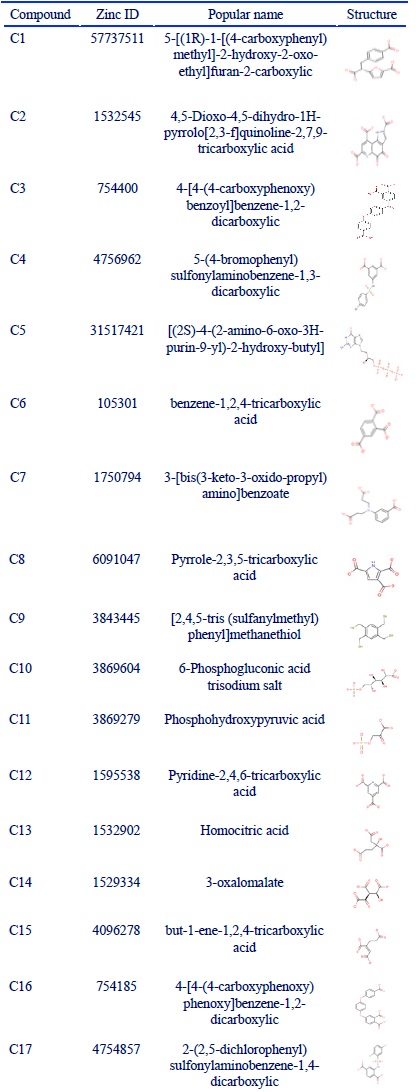
In order to evaluate TMLA cytotoxicity, MTT assay was utilized. HUVEC and A172 cell lines weretreated with increasing doses of TMLA. As shown in (Fig. 1), cell viability was decreased after exposure to TMLA in a dose-dependent manner. The IC50 was calculated at 13 mM concentration in HUVEC cell line. The inhibition ratio of this concentration was 30% in A172 cell line. With a dose-dependent increasing manner in A172 (14 and 15 mM), the cell viability was still around 62%.
Figure 1 :Cell Viability, Treatment of Huvec and A172 Cells with Different Concentration of Trimellitic Acid (Tmla). A: HUVEC cells were treated with TMLA at 8 concentrations of 1, 3, 5, 11, 12, 13, 14 and 15 mM for 24h. Cell viability was evaluated with MTT assay. IC50 was calculated 13 mM using Graph Pad instant 3.0 application. B: The effect of TMLA was evaluated at 12, 13, 14, 15 mM concentrations on A172 cell line (one way ANNOVA was used and p value of ***P<0.0001; **P<0.001; *P<0.05 were considered statistically significant comparing with control). The bars represent standard error of the mean of three replicates.
Real time PCR
Evaluation of gene expression was performed using real time PCR to analyze the inhibitory potential of TMLA on CD 31 expression as an endothelial cell marker. We compared gene expression of each concentration with 2 controls; 1- vitamin C as positive control and 2- free TMLA medium as negative control.Gene expression at 14, 13, 12, 0.5, and 0.1 mM concentration was dramatically decreased comparing with negative control (0.357, 0.307, 362, 0.282, and 0.285, respectively). Interestingly, gene expression of these concentrations were lower than positive control significantly (***P=0.0001). Gene expression among these concentrations was not significantly different (P>0.05)( Fig.2).
Figure 2 :Evaluation of Gene Expression of Cd31 Using Real Time Pcr. Vitamin C (Vit C) And Free Tmla Medium Were Utilized As Positive And Negative Controls Respectively. CD31 gene expression reduction of all concentrations was observed comparing with controls (***P<0.0001). No significant (P>0.05) difference between these concentrations (0.1-14 mM) was observed (one way ANNOVA was used and p value of ***P<0.0001; **P<0.001; *P<0.05 were considered statistically significant). The bars represent standard error of the mean of three replicates.
Tube Formation Assay
Tube formation is a good model for the investigation of angiogenesis inhibition. In this process, sprouting and migration of endothelial cells lead to establish cell-cell connections and are able to make polygon structures. Finally, these polygons can turn into tubules and form capillary like structure in vitro. The qualitative assay was carried out to evaluate the inhibitory effects of TMLA. VEGF and PBS were utilized as positive and negative controls, respectively. The result showed that VEGF stimulated formation of capillary tube while TMLA inhibited HUVEC tube formation at IC50 concentration (Fig.3)
Figure 3 : Qualitative Tube Formation Assay on Huvec Cell Line. Huvec cell line was treated with Vegf, Pbs, and TmlA. A: VEGF treatment as positive control induced formation of capillary tube; B: PBS treatment as negative control; C: TMLA treatment at IC50 (13 mM) inhibited tube formation.
Flow Cytometric Analysis
Staining with Annexin V-FITC and PI may distinguish between viable cells (negative for both annexin V-FITC and PI), early apoptosis (annexin V-FITC positive, PI negative), late apoptosis (annexin V-FITC positive, PI positive) or necrotic cells (annexin V-FITC negative, PI positive). In flow cytometry analysis of Annexin V-FITC/ PI double staining, the live cells, early apoptotic cells, late apoptotic cells, and necrotic cells were visible in the lower left (LL), lower right (LR), upper right (UR), and in the upper left (UL) quadrants, respectively.
In untreated HUVEC cells (control), 86.34% of the cells were viable, 2.63% was in early apoptosis, and 4.29% cells were in late apoptosis stage. When the HUVEC cells were treated with 5 mM, 10 mM, and 13 mM of the TMLA for 24 h, the viable cells were 76.35%, 71.32%, and 41.86%; the necrotic cells were 8.14%, 9.01%, and 12.49%; the early apoptotic cells were 1.95%, 0.5%, and 1.27%; and the late apoptotic cells were 13.56%, 19.13%, and 44.39%, respectively. All these data are shown in (Fig. 4). The results indicated that the effects of TMLA on HUVECs were through apoptosis in a dose dependent manner.
Figure 4 :Apoptosis Detection Using Flow Cytometry Analysis of Annexin V-Fitc/Pi
Huvecs were treated with different concentrations (5, 10 And 13 Mm) A: The numbers in the lower left considered to be viable cells (annexin V-/ PI-), while the upper left represented the percentage of necrotic cells (annexin V-/ PI+), in the lower right quadrant cells were considered to be in early apoptotic stage (annexin V+/ PI-), while the upper right represented the percentage of late apoptotic cells (annexin V+/PI-). B: Proportions of cells in the different subpopulations. The results indicated that the inhibition effects of TMLA on HUVECs were through apoptosis in a dose dependent manner. Statistical analysis was performed with one way ANNOVA and p value of ***P<0.0001 and **P<0.001 were considered statistically significant. The bars represent standard error of the means of three replicates.
Discussion
There are two classes of compounds which have the capacity to represent angiostatic activity via interruption of the ternary complex of FGF2- Heparin- FGFR1, including polycationic compounds that mimic positive charges of HBD and polyanionic compounds that compete with heparin to interact with heparin binding growth factors. Many substances and peptides were synthesized based on these two classes of compounds. Suramin [28], pentosan polysulfate [29], and bisphosphonates [30] are some of them that mimic heparin. Surfen [31] and porphyrin analogue [32] are positively charged agents that interact with heparin. There are also antiangiogenic agents based on natural inhibitors such as sm27 [33] and bi-naphthalenic compounds [34] that mimic FGF2 binding sequence of thrombospondin-1[35] and NSC12 based on pharmacophore modeling of Pentraxin 3-FGF2 interaction [36].
Rose et al., (2011) indicated that interaction of some bisphosphonates like clodronate and etidronate with FGF2 is probably at ATP binding domain [25]. Furthermore, the addition of foscarnet (with only one phosphonate group) to FGF2 at higher concentration compared to the bisphosphonates, reduces ATP-binding capacity to FGF2 [24].
Comparing these antiangiogenic agents (etidronate, clodronate, foscarnet, and fosfonet) indicated that 4.87 and 2.49 are common distances in foscarnet and fosfonet and common distances in clodronate and etidronate are 4.75 and 2.97, respectively. Intramolecular distances of ~4.7 and ~2.5 can be seen in ten and six molecules out of seventeen molecules we found in Zinc database respectively. Five of seventeen molecules had these two distances simultaneously (C5, C6, C8, C15, and C16). Among these five molecules, we chose TMLA due to availability and lack of toxicity. The considering distances for TMLA were 2.48 and 4.85.
In this study, we performed a detailed conformational analysis of the FGF2–Heparin and FGF2-Heparin-FGFR1 co-crystal structures and identified conformational characteristic of the heparin and the heparin-binding site on FGF2 that govern their interactions. A heparin that binds to both FGF2 and FGFR1 acts as a stimulator and a heparin that only binds to FGF2 acts as an inhibitor of signaling by sequestering the growth factor [21, 22]. Therefore, the difference regarding heparin interactions in FGF2-Heparin and FGF2-Heparin-FGFR1 complexes is important. Comparing the negative charge distances of heparin between both complexes of FGF2-Heparin and FGF2-Heparin-FGFR indicated that some of distances between sulfurs of heparin in FGF2-Heparin were different from those between sulfurs of FGF2-Heparin-FGFR (Table 2). For example, distances of 7.65, 7.42, and 7.35 between sulfurs of heparin in Heparin-FGF2 were different from those between sulfurs of heparin in FGF2-Heparin-FGFR1 that were 10, 4.75, and 4.61 respectively. This obvious difference suggested that the mentioned distances were probably important in the mode of interaction, that is the interaction of heparin with ligand or both ligand and receptor.
The numbers of 7.55, 7.41, and 7.30 were intramolecular negative charge distances of the chosen molecule, TMLA (C6), that are approximately the same as those possible important numbers of heparin that were in binding with just FGF2 (Table 4B)( Fig.5).
Figure 5 :Table 4B): Continued B
Furthermore, positive charge distances between K135 and R120 and between A136 and FGF2 in Heparin-FGF2 complex were 7.64 and 7.65, respectively; whereas, positive charge distances between those amino acids were obviously different in the presence of receptor (Table 1). As the residue K135 is essential for heparin-binding and it is in direct contact with sulfur anions of heparin [23], presumably sulfur anions of heparin with distance of 7.65 interact with positive charges, bridging between K135 and R120 or A136.
Considering HBD and ATP binding domains simultaneously, TMLA was chosen. Here, we demonstrated that TMLA had a concentration dependent influence on cell viability of HUVECs. Interestingly, no significant toxic effect was observed within the tested dose range of TMLA on A172 cell line with low FGFR expression. Accordingly, it seems that the inhibitory effect of TMLA is specific for FGF2 and does not rely on its cell toxicity. Gene expression was significantly decreased in comparison to negative and positive controls (p<0.05) and no significant difference was found between gene expressions of those concentrations (p>0.05). Thus, TMLA at concentration of 0.1 mM could induce responses equivalent to those achieved by concentrations up to 15 mM, indicating the saturation of the available FGF2 by 0.1 mM TMLA. Tube formation assay showed that presumed mode of action of TMLA after binding to FGF2 was to hinder the receptor activation by FGF2. To clarify whether the growth inhibitory effect of TMLA was related to the induction of apoptosis, we utilized flow cytometry assay. Increasing the numbers of apoptotic cells in a dose dependent manner (Figure 4) may prove that inhibitory effect of TMLA was through apoptosis.
Regardless of ATP binding domain of FGF2, molecular modeling studies clearly defined the differences regarding heparin interactions in FGF2-Heparin and FGF2-Heparin-FGFR1 by measuring distances of heparin anions and amino acids cations in both complexes. According to Table 2, other important distances may be around 13.6, 11.2 and 10.5 in FGF2-Heparin complex that were obviously different from the same distances in FGF2-Heparin-FGFR1 complex (15.17, 12.6 and 13Å). Therefore, those positive charged amino acids positioned in FGF2-Heparin complex similar to distances (13.6, 11.2 and 10.5) of sulfur anions of heparin in this complex were possibly involved in the interaction. For example, T121 and K135 interacted with sulfur groups of heparin. Similarly, there was a correspondence between R120 and K125 and between R120 and A136 in terms of their interaction with the sulfur anions of heparin. Thus, we hypothesized that a compound containing negative charge distances of ~ 13.6, 11.2, 10.5, and 7.6 may have high affinity and specificity for FGF2 binding. On the basis of our findings, among seventeen agents mentioned in Table 4, C1, C4, C7, and C17 were compounds that had this characteristic to some extent and offered the great opportunity for binding to FGF2. However, in vitro methods are clearly warranted to evaluate the potential role of them in inhibition of FGF2.
We are currently planning our next research step to inhibit FGFR1 through blocking HBD by designing highly specific agents. However, in our opinion, combination of FGFR1 inhibitor with TMLA to target FGF2 induced angiogenesis should be analyzed in the future. Based on defined interactions, our study identified a structural framework to design biomolecules with specific binding affinities to FGF2. The present work also showed that extension of FGF2 inhibitors with much specificity could be achieved. Considering the importance of their distances, it may be possible to make further derivatives of the agents with low molecular weight and much smaller and improve their properties while maintaining FGF2 inhibitory properties.
Acknowledgements
We thank Ghazaleh Ghavami for her technical advice on flow cytometry. This work was supported by Drug Design and Bioinformatics Unit, Medical Biotechnology Department, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran.
Conflict of interest
The authors declare no conflict of interest.
References
[1]. Ferrara N. Vascular endothelial growth factor as a target for anticancer therapy. oncologist. 2004;volume 9(Supplement 1):2-10.
[2]. Folkman J. What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst. 1990; 82(1):4-7.
[3]. Herbst RS. Therapeutic options to target angiogenesis in human malignancies. Expert Opin Emerg Drugs. 2006;11(4):635-50.
[4]. Garcia-Fernandez L, Halstenberg S, Unger RE, Aguilar MR, Kirkpatrick CJ, San Roman J. Anti-angiogenic activity of heparin-like polysulfonated polymeric drugs in 3D human cell culture. Biomaterials. 2010;31(31):7863-72.
[5]. Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2(3):1-12.
[6]. Nugent MA, Iozzo RV. Fibroblast growth factor-2. Int J Biochem Cell Biol. 2000;32(2):115-20.
[7]. Bikfalvi A, Klein S, Pintucci G, Rifkin DB. Biological roles of fibroblast growth factor-2. Endocrine reviews. 1997;18(1):26-45.
[8]. Kan M, Wang F, Xu J, Crabb JW, Hou J, McKeehan WL. An essential heparin-binding domain in the fibroblast growth factor receptor kinase. Science. 1993;259(5103):1918-21.
[9]. Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285(21):1182-6.
[10]. Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med. 1995;1(1):27-31.
[11]. Casu B, Lindahl U. Structure and biological interactions of heparin and heparan sulfate. Adv Carbohydr Chem Biochem. 2001;57:159-206.
[12]. Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729-77.
[13]. Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem. 1998;67:609-52.
[14]. Fernandez-Tornero C, Lozano RM, Redondo-Horcajo M, Gomez AM, Lopez JC, Quesada E, et al. Leads for development of new naphthalenesulfonate derivatives with enhanced antiangiogenic activity: crystal structure of acidic fibroblast growth factor in complex with 5-amino-2-naphthalene sulfonate. J Biol Chem. 2003;278(24):21774-81.
[15]. Lindahl U, Lidholt K, Spillmann D, Kjellén L. More to “heparin” than anticoagulation. Thromb Res. 1994;75(1):1-32.
[16].Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem. 1998;67:609-52.
[17]. Klumpp S, Kriha D, Bechmann G, Maaßen A, Maier S, Pallast S, et al. Phosphorylation of the growth factors bFGF, NGF and BDNF: a prerequisite for their biological activity. Neurochem Int. 2006;48(2):131-7.
[18]. Thompson LD, Pantoliano MW, Springer BA. Energetic characterization of the basic fibroblast growth factor-heparin interaction: identification of the heparin binding domain. Biochemistry. 1994;33(13):3831-40.
[19]. Li L-Y, Safran M, Aviezer D, Boehlen P, Seddon AP, Yayon A. Diminished heparin binding of a basic fibroblast growth factor mutant is associated with reduced receptor binding, mitogenesis, plasminogen activator induction, and in vitro angiogenesis. Biochemistry. 1994; 33(36):10999-1007.
[20]. Rose K, Gast RE, Seeger A, Krieglstein J, Klumpp S. ATP-dependent stabilization and protection of fibroblast growth factor 2. J Biotechnol. 2010;145(1):54-9.
[21]. Friedl A, Chang Z, Tierney A, Rapraeger AC. Differential binding of fibroblast growth factor-2 and -7 to basement membrane heparan sulfate: comparison of normal and abnormal human tissues. Am J Pathol. 1997;150(4):1443-55.
[22]. Chang Z, Meyer K, Rapraeger AC, Friedl A. Differential ability of heparan sulfate proteoglycans to assemble the fibroblast growth factor receptor complex in situ. FASEB J. 2000;14(1):137-44.
[23]. Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6(3):743-50.
[24]. Rose K. Foscarnet reduces FGF2-induced proliferation of human umbilical vein endothelial cells and has antineoplastic activity against human anaplastic thyroid carcinoma cells. Biomed Pharmacother. 2013;67(1):53-7.
[25]. Rose K, Finger IE, Ferenz KB. Interaction of clodronate with fibroblast growth factor 2 reduces FGF2-activity in endothelial cells. Biomed Pharmacother. 2011;65(1):46-51.
[26]. Mikirova NA, Ichim TE, Riordan NH. Anti-angiogenic effect of high doses of ascorbic acid. J Transl Med. 2008;6:50.
[27]. Horak E, Klenk N, Leek R, LeJeune S, Smith K, Stuart N, et al. Angiogenesis, assessed by platelet/ endothelial cell adhesion molecule antibodies, as indicator of node metastases and survival in breast cancer. Lancet. 1992; 340(8828):1120-4.
[28]. Gagliardi A, Hadd H, Collins DC. Inhibition of angiogenesis by suramin. Cancer Res. 1992;52(18):5073-5.
[29]. Nguyen NM, Lehr JE, Pienta KJ. Pentosan inhibits angiogenesis in vitro and suppresses prostate tumor growth in vivo. Anticancer Res. 1993;13(6A):2143-7.
[30]. Ziebart T, Pabst A, Klein MO, Kammerer P, Gauss L, Brullmann D, et al. Bisphosphonates: restrictions for vasculogenesis and angiogenesis: inhibition of cell function of endothelial progenitor cells and mature endothelial cells in vitro. Clin Oral Investig. 2011;15(1):105-11.
[31]. Schuksz M, Fuster MM, Brown JR, Crawford BE, Ditto DP, Lawrence R, et al. Surfen, a small molecule antagonist of heparan sulfate. Proc Natl Acad Sci U S A. 2008;105(35):13075-80.
[32]. Aviezer D, Cotton S, David M, Segev A, Khaselev N, Galili N, et al. Porphyrin analogues as novel antagonists of fibroblast growth factor and vascular endothelial growth factor receptor binding that inhibit endothelial cell proliferation, tumor progression, and metastasis. Cancer Res. 2000;60(11):2973-80.
[33]. Colombo G, Margosio B, Ragona L, Neves M, Bonifacio S, Annis DS, et al. Non-peptidic thrombospondin-1 mimics as fibroblast growth factor-2 inhibitors: an integrated strategy for the development of new antiangiogenic compounds. J Biol Chem. 2010;285(12):8733-42.
[34]. Foglieni C, Pagano K, Lessi M, Bugatti A, Moroni E, Pinessi D, et al. Integrating computational and chemical biology tools in the discovery of antiangiogenic small molecule ligands of FGF2 derived from endogenous inhibitors. Sci Rep. 2016;6:23432.
[35]. Manetti F, Corelli F, Botta M. Fibroblast growth factors and their inhibitors. Curr Pharm Des. 2000;6(18):1897-924.
[36]. Ronca R, Giacomini A, Di Salle E, Coltrini D, Pagano K Ragona L, et al. Long-Pentraxin 3 Derivative as a Small-Molecule FGF Trap for Cancer Therapy. Cancer Cell.2015;28(2):225-39.
License
Copyright
© ,
Author Details