High-level Chemoresistance in Paclitaxel-resistant C-33A Cells Involves the Alteration of Multiple Signaling Pathways and the Drug Efflux Phenotype by Modulating Coding and Long Non-coding RNAs
Download
Abstract
Background: In chemoresistance, a variety of oncogenic signaling pathways can be activated and interconnected to combat drug toxicity, making anticancer pharmacological strategies complex.
Objective: This study aimed to generate a paclitaxel-chemoresistant cancer cell line scaffold and explore its mechanisms of chemoresistance.
Materials and methods: Total RNAs from C-33A and C-33A RPTX (paclitaxel-resistant) cells were sequenced to determine the transcriptome of coding and long non-coding RNAs (lncRNAs). Enrichment analysis of differentially expressed genes (DEGs) was conducted. Flow cytometry was performed to analyze doxorubicin accumulation in C-33A RPTX cells.
Results: C33-A RPTX cells achieved a resistance index of 55. 1332 genes were differentially expressed (log2 fold-change>1). Gene ontology (GO) analysis revealed that biological processes, such as angiogenesis and mesenchymal development, were altered, while the mainly altered cellular components were from the extracellular matrix (ECM). Meanwhile, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that signaling pathways, including PI3K/Akt, MAPK, Calcium, ECM-receptor, Rap1, EGFR tyrosine kinase inhibitor resistance, tight junction, and platinum drug resistance, were altered by coding RNAs or long non-coding RNAs (lncRNAs). Finally, the overexpression of the ABCB1 gene in C-33A RPTX cells correlated with a drug efflux phenotype, as evidenced by reduced cellular accumulation of the reporter drug doxorubicin. Alterations in signaling pathways recognized as relevant to chemoresistance and increased drug efflux, driven by changes in the expression patterns of coding RNAs and lncRNAs, may be cooperating to establish resistance to paclitaxel in C-33A RPTX cells.
Conclusions: This C-33A RPTX model provides a new experimental platform to study the molecular mechanisms of chemoresistance in cervical cancer. The study integrates both coding and non-coding RNAs, revealing their coordinated alteration in chemoresistance-related signaling pathways. Thus, this dual-level analysis represents a novel systems-level insight into paclitaxel resistance. Beyond descriptive transcriptomics, this resistant model is proposed as a biological platform for testing novel antineoplastic compounds and exploring their mechanisms of action, giving it translational application.
Introduction
Cancer is one of the leading causes of death worldwide, with 9.7 million deaths in 2022 [1]. Approximately 80- 90% of cancer deaths are attributed to chemoresistance [2, 3]. Tumor heterogeneity is thought to be the main obstacle in cancer treatment due to the existence of intrinsic or acquired chemoresistance clonal lineages. Chemoresistance leads to cancer recurrence that eventually causes therapeutic failure and death of cancer patients [2].
Paclitaxel is one of the most widely used antineoplastic agents across a range of cancer types due to its diverse pharmacological targets and mechanism of action [4]. The canonical mechanism of action of paclitaxel is mitotic arrest by promoting microtubule polymerization and stabilization [5]. However, there is evidence that other non-antimitotic mechanisms may be more decisive in achieving the cytotoxic effect as an anticancer agent. Paclitaxel-induced multiple micronuclei formation is a cytotoxic mechanism independent of microtubule binding and mitotic arrest. The formation of various nuclei results from the rupture of the nuclear envelope, which occurs in malignant cells but not in normal cells. It is associated with a reduction in the nuclear lamina protein lamin A/C [6, 7]. Ferlini and colleagues have shown that paclitaxel binds directly to the antiapoptotic Bcl-2 in a remarkably similar manner to tubulin and mimics the activity of nur77, transforming the antiapoptotic activity of Bcl-2 to a proapoptotic one [8].
However, resistance to paclitaxel appears to halt efficacy in chemotherapy. A variety of paclitaxel resistance mechanisms have been addressed in different types of cancer. One of the most studied resistance mechanisms that mitigates the cytotoxic effect of paclitaxel and many other drugs is the overactivation of drug efflux through the overexpression of ATP-binding cassette (ABC) membrane transporters, which results in decreased intracellular drug concentrations [9]. Among the members of this extensive family of transporters, the ABCB1 gene encoding P-glycoprotein (P-gp) is clinically the most relevant due to its presence in virtually all types of cancer, including cervical [10], ovarian [11], breast [12], gastric [13], lung [14], and liver cancers [15]. A resistance mechanism that concerns antimitotic antineoplastics such as paclitaxel is the isotype switch of β-tubulin. For decades, studies have shown that cell lines of different types of cancer resistant to paclitaxel present an alteration in the content and isotype of β-tubulin, among which the βIII- and βIV- tubulin isotypes stand out [16].
Additionally, the mechanisms of resistance to paclitaxel have been explored at other levels of gene expression regulation, including microRNAs and lncRNAs. LncRNA H19 has been associated with chemoresistance in breast cancer [17]. A study by Jiguang et al. (2018) has shown the relationship between high levels of lncRNA H19 expression and resistance in paclitaxel-resistant cell lines, while H19 knockdown recovered their chemosensitivity to paclitaxel [18].
For instance, Dragicevic et al. (2025) performed a comprehensive RNA-seq analysis in colon cancer cells and identified DEGs related to metabolism, drug efflux, apoptosis, and extracellular matrix organization, revealing that the PI3K-Akt and MAPK pathways were strongly associated with paclitaxel resistance [19]. Similarly, Sarkar et al. (2025) from transcriptomic analysis of basal-like breast cancer identified KHDRBS3 protein as a key gene associated with paclitaxel resistance and stemness properties. Elevated KHDRBS3 expression correlated with poor overall survival and reduced response to taxane-based therapy [20].
As knowledge emerges around the molecular mechanisms underlying cancer chemoresistance, the complexity and interrelation of various cell signaling pathways are demonstrated, where cell type seems to play a determining role.
In the present study, we generated a high-level laboratory paclitaxel-resistant cervical cancer cell line, C-33A RPTX. Because numerous studies report that paclitaxel-resistant cancer cells are established, at least in part, by a drug efflux phenotype, we characterized the transcriptomic profile of C-33A RPTX cells to identify the regulation of coding and long non-coding RNAs, which could lead to the ABCB1-mediated drug efflux phenotype.
Materials and Methods
Cervical Cancer Cell Line Culture
The C-33A cell line derived from cervical cancer was kindly provided by Alejandro García-Carrancá (IIB, UNAM, Ciudad de México, México). It was cultured in Dulbecco’s Modified Eagle’s Medium High Glucose (4.5 g/L) with L-glutamine and sodium pyruvate, supplemented with Fetal Bovine Serum (10% v/v) and penicillin-streptomycin (71% v/v, 100 U/mL final), at 37 C̊ and 5% CO2.
Cell Viability Assay
To determine the mean inhibitory concentration (IC 50, the concentration of paclitaxel that reduces cell viability by 50%) of paclitaxel in C-33A cells, 6x103 cells were seeded per well in a 96-well plate. After overnight, cells were exposed to 0, 2.5, 5, 10, 20, 40, 80, and 100 nM of paclitaxel during 72 h. Dimethyl sulfoxide was used as a vehicle. Six treatment replicates were made for each concentration, then, cell viability was measured using the colorimetric method [3-(4,5-dimethylthiazol-2-yl)- 5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H- tetrazolium] inner salt MTS assay (Promega, Madison, WI, USA), following the manufacturer’s instructions. The absorbance values at 492 nm were recorded in a microplate reader using the Bichromatic program. The cell viability percentages relative to the control (vehicle) were calculated, and the IC50 value was determined by non-linear regression analysis using the method [inhibitor] vs normalized response in GraphPad PRISM.
Generation of Resistance to Paclitaxel in the C-33A Cell Line
To generate a paclitaxel-resistant cancer cell line, a subconfluent plate of C-33A cells was exposed to a sublethal concentration of paclitaxel, 4.5 nM, for 72 h. Then, cells were trypsinized and maintained in free-paclitaxel media culture for 96 h for recovery. One passage was made between treatments and recoveries. Three cycles of treatment and recovery were made with each employed concentration. Progressive increases of paclitaxel of 9, 18, and 36 nM were carried out.
Determination of Chemoresistance to Paclitaxel in C-33A Cells
After chronic exposure to paclitaxel, the level of resistance to the drug was determined in the C-33A RPTX cell line. For that, the cell viability of C-33A RPTX cells and the IC50 against PTX were determined as in the point “2.2 Cell Viability Assay,” except that the concentrations of exposure were 7.8, 15.6, 31.2, 62.5, 125, 250, 500, 1000, and 2000 nM PTX.
RNA extraction and quality of total RNA of C-33A and C-33A RPTX cell lines
Total RNA was isolated from C-33A and C-33A RPTX cell lines with TRIzol in a subconfluent 60 mm cell culture plate. The RNA concentration was evaluated with the NanoDrop 2000 Spectrophotometer, and the RNA Quality was evaluated by an Agilent 2100 Bioanalyzer; samples with RIN >8 were sequenced.
RNA sequencing of C-33A and C-33A RPTX cell lines
rRNA was depleted using Illumina TruSeq stranded total RNA with the Ribo-Zero gold kit according to the manufacturer’s protocol. The cDNA was constructed from fragmented RNA using TruSeq RNA Sample Prep v2 Kits. The library was sequenced by Novogene Co, Ltd. using the Illumina NovaSeq 6000 platform.
Quantification of gene expression level
To quantify gene expression levels, FPKM was calculated from the count of reads assigned to each gene using featureCounts v1.5.0-p3. Differential expression analysis between C-33A and C-33A RPTX cells was conducted using the DESeq2 R package Version 1.20.0. P-values were adjusted using the Benjamini and Hochberg method. A corrected P-value of 0.05 and absolute log2 foldchange ≥1 were set as the threshold for significantly differential expression.
Enrichment analysis of differentially expressed genes
The clusterProfiler R package (version 3.8.1) was used to calculate corrected p-values for the DEGs. GO terms with a corrected p-value less than 0.05 were considered significantly enriched DEGs. The Clusterprofiler R package 3.8.1 was also used to test the statistical enrichment of DEGs in KEGG pathways and GO terms analysis. Additionally, NcPath was used for enrichment analysis of lncRNAs on KEGG pathways.
Doxorubicin cell accumulation
3x105 C-33A or C-33A RPTX cells were cultured per well in 6-well plates. After overnight, the cells were treated with 0.1 µM doxorubicin for 2 h. Subsequently, the cells were washed with PBS and collected by trypsin 0.25 %. The cells were then centrifuged at 1000 rpm for 3 min and washed with PBS. Cells were resuspended in 1 mL of PBS and analyzed by flow cytometry. Ten thousand events were acquired for each sample. The histograms were made and analyzed by FlowJo software to compare the doxorubicin cell accumulation in both C-33A and C-33A RPTX cells.
Statistical Analysis
Statistical significance of the data was performed in GraphPad Prism with a one-way ANOVA with a 95% confidence interval, followed by a Dunnett test to compare each group against the control.
Results
Assessment of the sensitivity of the C-33A cervical cancer cell line to paclitaxel
To determine the sensitivity of the C-33A cell line to paclitaxel, the cells were exposed to different concentrations of paclitaxel for 72 h. After exposure, the IC50 was determined. Figure 1 shows the IC50 of paclitaxel in C-33A cells, which was 11.78 nM.
Figure 1. Cell Viability Curve of C-33A Cells Treated with Paclitaxel. Cells were treated with 2.5, 5.0, 10, 20, 40, 80, and 100 nM of paclitaxel for 72 hours. The mean and error bars representing the percentage of cell viability are shown. * p<0.05; **** p<0.0001 .
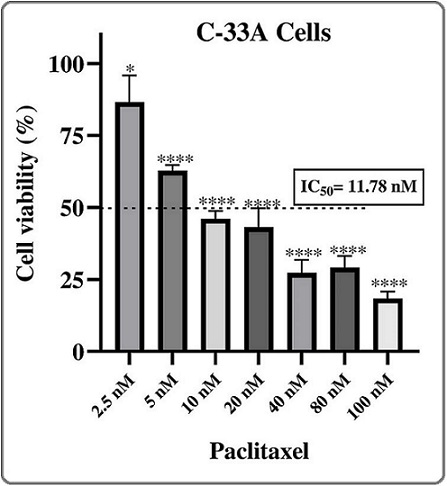
The IC50 was used as a reference to initiate chronic exposure to a sublethal concentration of paclitaxel in order to generate chemoresistant C-33A cells.
Paclitaxel induces elevated chemoresistance in C-33A cervical cancer cells
After chronic exposure to a sublethal concentration of paclitaxel of 4.5 nM and subsequent gradual increase of the concentration to 36 nM, the cell viability of C-33A cells was assessed in a concentration range of 7.8 to 2000 nM of paclitaxel. The results showed that C-33A cells (named from now on C-33A RPTX) achieved a notable chemoresistance to paclitaxel, with an IC50 of 655.2 nM (Figure 2), which corresponded to a resistance index (RI) of 55.
Figure 2. Cell Viability Curve of C-33A RPTX Cells Treated with Paclitaxel. Treatments with 7.8, 15.6, 31.2, 62.5, 125, 250, 500, 1000, and 2000 nM of paclitaxel for 72 h were employed. The mean and error bars of the percentage of cell viability were plotted. * p<0.05; ** p<0.01; **** p<0.0001 .
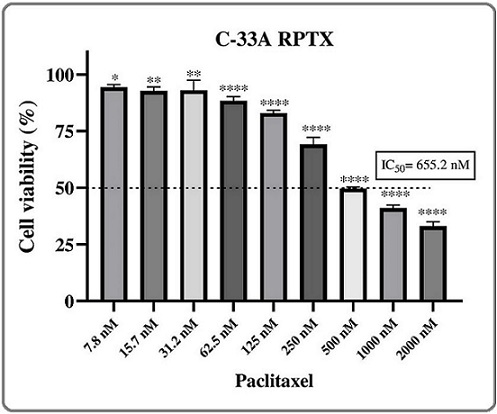
That represented that C-33A RPTX cells required a 55-fold paclitaxel concentration to achieve the same effect as C-33A parental cells.
Classification of differentially expressed genes between C-33A and C-33A RPTX cells
The entire transcriptomics of C-33A and C-33A RPTX cells were analyzed with the objective of identifying differences that indicate the causes of chemoresistance to paclitaxel. Initially, we discovered that 1,332 genes were differentially expressed with a log2 fold-change >|1|, of which 864 genes were upregulated, and 468 genes were downregulated (Figure 3).
Figure 3. Differential Expression Analysis and Gene Classification of Transcriptomics in C-33A and C-33A RPTX Cells. Volcano plot graphs showing upregulated (864) and downregulated (468) genes (A). Bar graph of 1332 classified DEGs (B); 1126 (85%) of all RNAs are coding genes, 153 (11.5%) are long non-coding RNAs, 35 (2.6%) are pseudogenes, 10 (0.7%) are unknown genes, and the remaining include a snRNA, snoRNA, and artifacts.
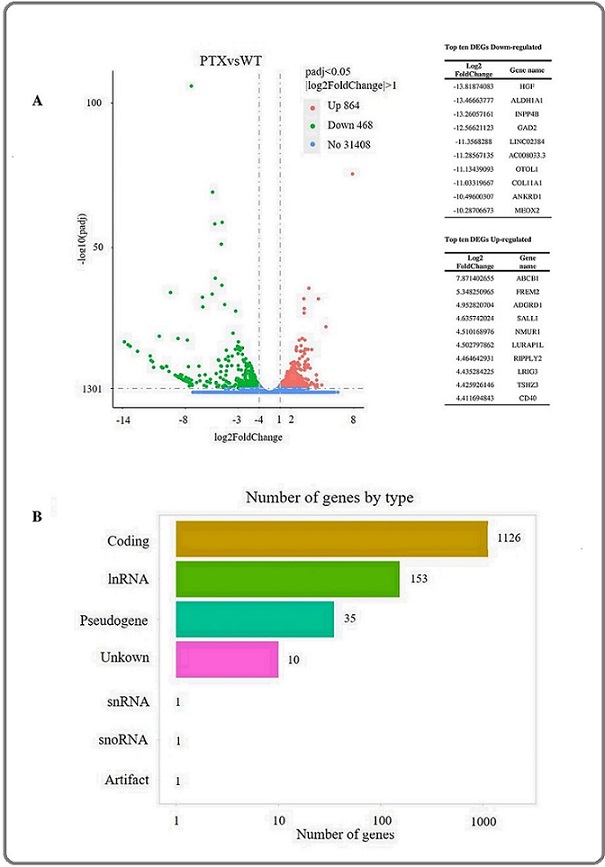
The gene type classification revealed that 85% of these were coding genes, 11.5% were long non-coding RNA (lncRNA) genes, and the remaining 3.5% consisted of pseudogenes, unknown genes, and small RNAs.
Gene ontology enrichment of differentially expressed genes between C-33A and C-33A RPTX cells
The GO analysis of the DEGs revealed that genes with a minor adjusted p-value (p < 0.05) were associated with biological processes such as angiogenesis, mesenchyme development, morphogenesis of a branching epithelium, and other biological processes related to the heart (Figure 4).
Figure 4. Gene Ontology Analysis of Differentially Expressed Genes between C-33A and C-33A RPTX Cells. Bar chart of the ten most significant enrichments (-log10 (padj)) of the Biological Processes (BP), Cellular Components (CC), and Molecular Functions (MF) of the DEGs. Numbers at the top of each chart represent the number of genes associated with that GO term.
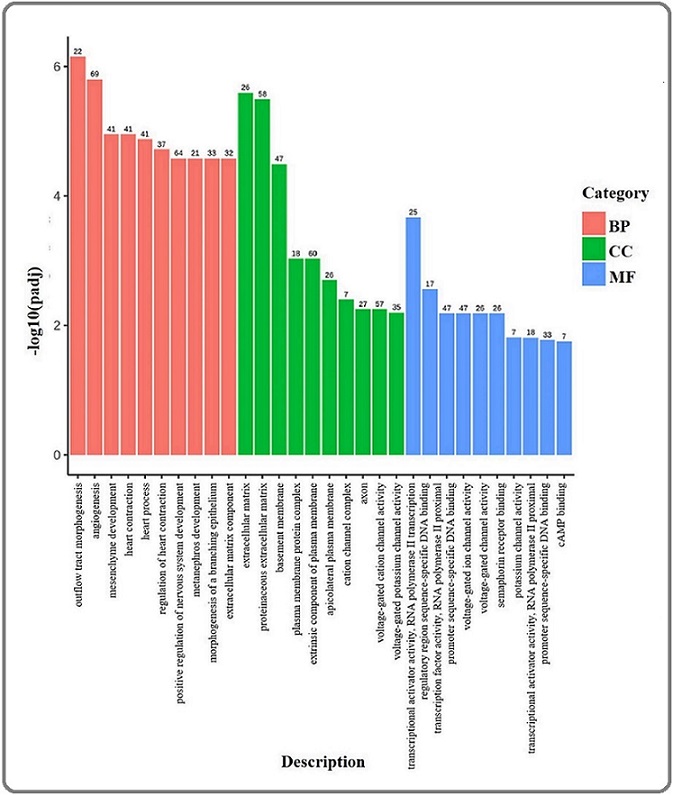
Meanwhile, the primary molecular functions associated with DEGs included ion transport channel activities, transcriptional activities, and other less prominent overrepresented functions (Table 1S). Regarding cellular components, the DEGs that were mainly overrepresented included extracellular matrix components, the extracellular matrix itself, the proteinaceous extracellular matrix, and the basement membrane, among others.
Metabolic pathways were differentially modulated between C-33A and C-33A RPTX cells
The KEGG pathway analysis revealed that coding RNAs differentially expressed in C-33A and C-33A RPTX cells with the minor padj value were mainly associated with several signaling pathways involved in cancer and chemoresistance, such as calcium signaling, MAPK signaling, focal adhesion, Rap1 signaling, PI3K- Akt signaling, cAMP signaling, ECM-Receptor, and Cell adhesion, and others (Figure 5a and Table 2S) that could promote chemoresistance to paclitaxel in C-33A RPTX cells.
Figure 5. Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis of Differentially Expressed Genes. Dot chart showing the most significant DEGs with adjusted p-values (padj), associated with metabolic pathways of coding RNA (A) and lncRNAs (B). The size of each dot corresponds to the GeneRatio (the ratio of differentially expressed genes to all genes for this GO term).
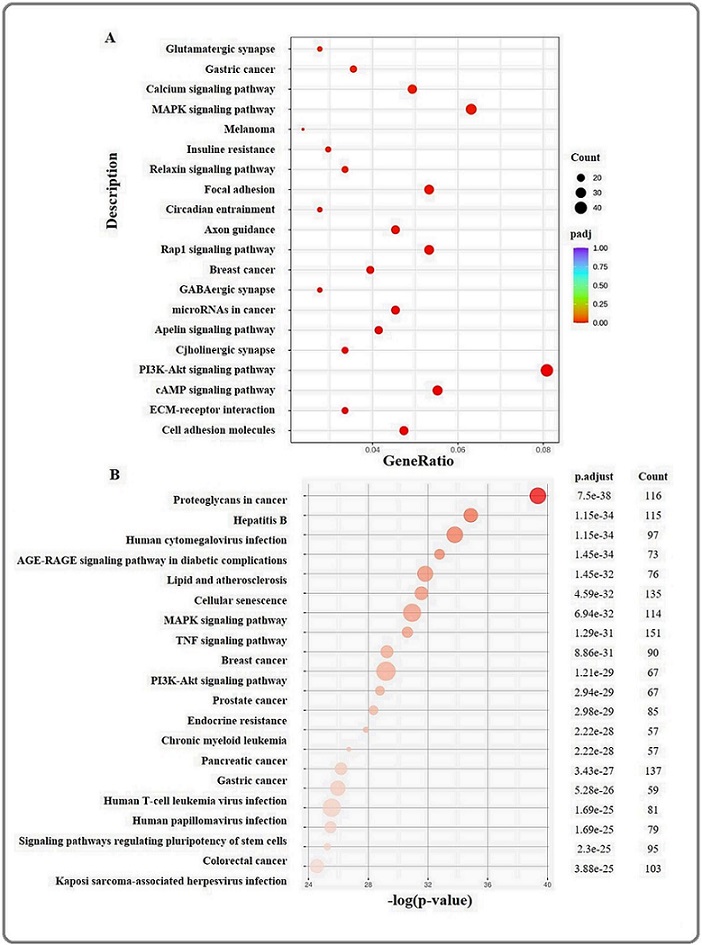
It is known that lncRNAs act as regulators of gene expression at different levels; therefore, a group of 153 lncRNAs differentially expressed between C33A and C-33A RPTX cells was analyzed for KEGG pathways enrichment. The results showed that those were associated as regulators of genes involved in 185 metabolic pathways, such as MAPK signaling, TNF signaling, PI3K-Akt signaling, EGFR tyrosine kinase inhibitor resistance, adherents junction, ErbB signaling, platinum drug resistance, apoptosis, and tight junction (Figure 5b and Table 3S), that could boost the chemoresistance of C-33A RPTX cells to paclitaxel. These data indicate that chemoresistance in C-33A RPTX cells involves the regulation of both coding and lncRNA expression.
A cluster of upregulated ATP-binding cassette transporter genes and a diminished doxorubicin cellular accumulation are present in C-33A RPTX cells.
Numerous reports have indicated that different types of paclitaxel-resistant cancer cells overexpressed ABC transporter genes, such as ABCB1. As a consequence, reduced intracellular drug accumulation is observed. In this study, a reduced ABC transporter gene group was differentially expressed (Table 4S). From the list of all DEGs between C-33A and C-33A RPTX cells, the ABCB1 gene was the most upregulated in C-33A RPTX cells. In order to know whether C-33A RPTX cells exhibited an intracellular drug concentration reducer phenotype, we carried out an exploratory assay at different times (1, 2, and 3 h) and concentrations (0.1 and 1 µM) of doxorubicin in both C-33A and C-33A RPTX cells. It was observed that in nearly all conditions, except for 0.1 µM doxorubicin applied for 1 h, C-33A RPTX cells demonstrated a significant intracellular drug reduction compared to C-33A cells (Figure 6).
Figure 6. Flow Cytometric Analysis of Doxorubicin Accumulation in C-33A and C-33A RPTX Cells. Histograms show flow cytometry data for C-33A (A) and C-33A RPTX cells (B) stained with 0.1 and 1 µM doxorubicin for 1, 2, and 3 hours. Controls consisted of C-33A or C-33A RPTX cells without doxorubicin staining. Doxorubicin fluorescence is shown on the X-axis, and the number of cells on the Y-axis. To the right, the figure displays the percentage of doxorubicin retention relative to cells without doxorubicin for each condition tested in both cell lines.
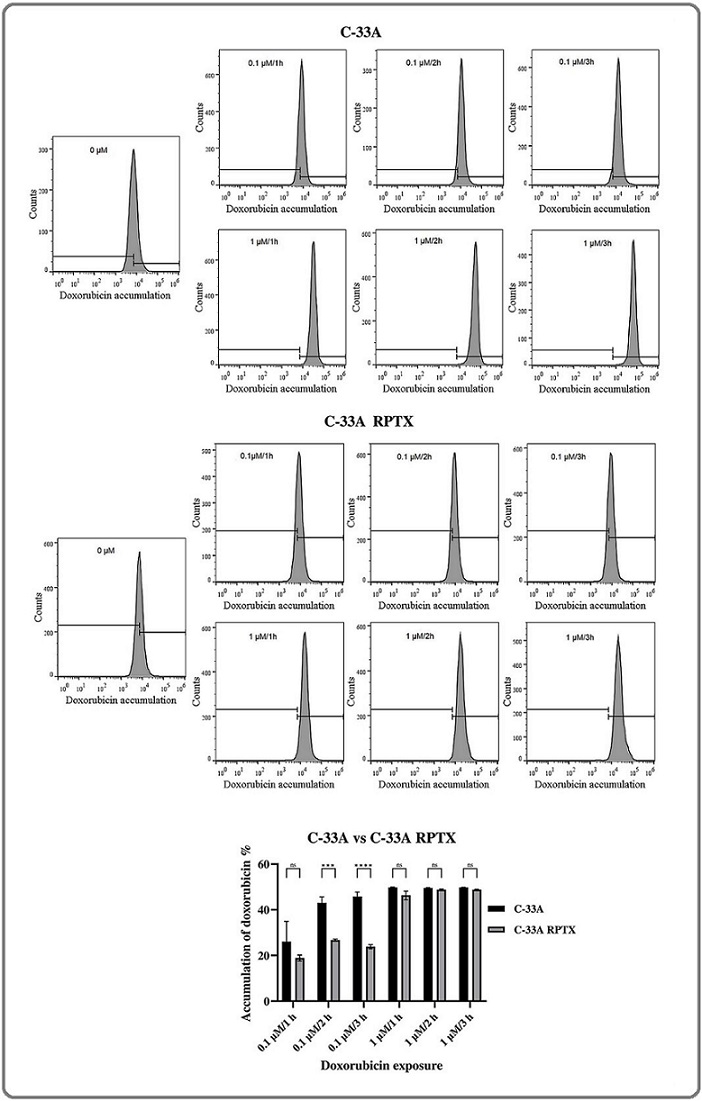
These results confirmed the drug efflux phenotype in C-33A RTPX cells.
Discussion
The use of monochemotherapy is generally not a highly effective strategy. However, combination chemotherapy is more effective and reduces side effects. The efficacy of paclitaxel in combination with platinum drugs has made the use of these drugs the first choice for advanced cervical cancer [21], ovarian cancer [22], advanced gastric cancer, and metastatic esophageal cancer [23]. Nevertheless, chemoresistance appears to depict a major barrier to the clinical efficacy of the treatment of cervical cancer [24]. The recurrence and persistence of the disease, driven by resistance to chemoradiotherapy, underlie the high mortality rates in cervical cancer patients. The overall survival in cervical cancer patients diagnosed with advanced-stage disease or metastasis is poor, with only a 1-year survival of 10–20% [25]. A diversity of resistance mechanisms to paclitaxel has been reported in several cancer cell types; those could be a consequence of each cancer cell type’s genetic and phenotypic backgrounds.
In this study, a paclitaxel-resistant cervical cancer cell line, C-33A RPTX cells, was generated through chronic exposure to the drug by progressive increases in the concentration. The RI achieved was 55-fold, relative to C-33A parental cells. According to McDermott [26], this level of chemoresistance corresponds to a High-level labotory model; se repite en un párrafo muy pequeño. This type of chemoresistance model provides a controlled system to identify molecular mechanisms of resistance at different levels of the regulation of gene expression, such as transcriptomics, proteomics, metabolomics, and so on [27, 28]. Also enables hypothesis-driven testing of interventions (inhibitors, drug combinations, genetic perturbations) in a platform where effects on resistant phenotypes are measurable and repeatable [29]. In addition, this model acts as a discovery engine for biomarkers of resistance that can be translated into clinical assays after validation [30, 31]. However, this model has limitations, such as the lack of clonal heterogeneity resulting from selection with drug exposure. Therefore, this model should be viewed as a bridge to more complex models that will support clinical translation.
The enrichment analysis of gene ontology showed that KEGG analysis associated DEGs (coding) with cancer and chemoresistance signaling pathways, such as calcium signaling, MAPK signaling, focal adhesion, Rap1 signaling, PI3K-Akt signaling, cAMP signaling, ECM-Receptor, and Cell adhesion.
Calcium signaling is a universal pathway that regulates several cellular processes, such as exocytosis, motility, gene transcription, muscle contraction, proliferation, and cell survival [32-34]. Shortt et al. (2012) described calcium as a key regulator of cell survival and apoptosis. Alterations in calcium signaling can disrupt the balance between pro-survival and pro-death pathways, allowing cancer cells to evade apoptosis. This dysregulation contributes to chemoresistance by enhancing cell survival mechanisms and reducing the effectiveness of cytotoxic drugs [35]. Ivanova et al. (2017) highlighted that calcium flux between the endoplasmic reticulum and mitochondria sustains cancer cell survival. By maintaining mitochondrial metabolism and preventing calcium overload–induced apoptosis, altered Ca²⁺ signaling supports resistance to chemotherapy. Thus, disrupted ER-mitochondrial calcium transfer is a key mechanism promoting chemoresistance in cancer cells [36]. For years, Kidd et al. (2002) reported that an early cytosolic effect of paclitaxel involved a rapid decrease in mitochondrial membrane potential and a loss of mitochondrial Ca2+, without significant effects on microtubule organization [37]. Likewise, the effect of several chemotherapeutic drugs, such as cisplatin, adriamycin, and ATO, is dependent on calcium transfer in mitochondria-associated endoplasmic reticulum membranes (MAMs) [38]. These studies show the relevance of calcium signaling in resistance to antineoplastic agents, such as PTX, and strengthen the hypothesis that in our study, the alteration of the an effect on the resistance of C-33A RPTX cells.
Meanwhile, the MAPK pathway comprises a group of serine/threonine kinases that can be aberrantly activated in cancer, playing an important role in cell survival, the development of tumors, and chemoresistance [39, 40]. Like MAPK, Akt is a serine/threonine kinase that promotes protein synthesis, proliferation, cell survival, and chemoresistance through the phosphorylation of many proteins. Akt is activated through phosphorylation by serine/threonine kinases via PI3K, which in turn is activated by membrane receptors. Furthermore, MAPKs and PI3K-Akt pathways are activated by the same membrane receptor, the FGFR [41]. In paclitaxel-resistant ovarian cancer cells, resistance was reversed by tephrosin treatment, which in turn decreased FGFR phosphorylation. Indeed, in that study, it was also found to decrease the active forms of the ERK and p38 MAPKs, as well as AKT. Uniquely, the combination, but not tephrosin or paclitaxel alone, induced apoptosis in paclitaxel-resistant ovarian cancer cells [42]. The study showed that different signaling pathways might be simultaneously involved in chemoresistance and that the sensitivity of paclitaxel- resistant cancer cells can be restored by inhibiting these signaling pathways, such as MAPKs and PI3-K/Akt. These previous findings could explain the alteration of genes of these PI3K/AKT and MAPKs signaling pathways in the transcriptomics of this study. On the other hand, the participation of p38 MAPK in the depolymerization of microtubules is well documented [43, 44]. Therefore, the aberrant activation of the MAPK signaling pathway enhances chemoresistance phenotype against microtubule- targeting agents (MTA), such as paclitaxel. Meanwhile, the inhibition of p38 MAPK enhances the efficacy of MTA [44]. These reports add significance to the findings of our transcriptomic study in which paclitaxel-resistant cells show the alteration of genes of the MAPK signaling pathway, pointing to the implication that these may have in the phenotype of our resistance model by counteracting the antimitotic effect of paclitaxel-induced polymerization and stabilization of microtubules.
Rap1 is a small GTPase that regulates several downstream signaling pathways, including MAPKs. Different stimuli regulating cell growth direct the signal through Rap1. Furthermore, Rap1 regulates different cellular processes such as cell adhesion, integrin activation, and cell-cell junction formation. Its participation in chemoresistance has also been widely reported [45-47]. In ovarian cancer, sensitivity to paclitaxel is dependent on the activity of the Rap1B isoform. This is supported by the depletion of the circular RNA, circ_0000231, which led to the inhibition of resistance, proliferation, invasion, migration, and epithelial-mesenchymal transition (EMT), and the induction of apoptosis, whereas overexpression of circ_0000231 led to the opposite effect. It was shown that circ_0000231 acted as a sponge for microRNA-140 (miR140), and Rap1B was a target of miR-140 [48]. These findings lead us to hypothesize that the Rap1 signaling pathway could have an effect on the resistance generated in C-33A RPTX cells.
In this work, gene ontology and KEGG pathway analysis revealed differential expression of genes related to biological processes, cellular components, and ECM signaling pathways in C-33A RPTX cells. The loss of cell-cell adhesion is crucial for cancer cells to invade other tissues. In this process, the compositional and signaling changes of the ECM are fundamental [49, 50]. In addition to the adhesion, migration, and invasion processes, which depend on ECM signaling, the resistance to chemotherapeutic drugs is also promoted by ECM. In principle, the modified composition of ECM proteins results in a physical barrier for drugs, decreasing drug effectiveness [51]. In turn, the changes in ECM composition also modify cell signaling transduction and produce chemoresistance through the activation of the EMT [52], MAPKs [53], and PI3K/Akt [53] pathways, including the overexpression of ABCB1 [53, 54].
In this work, the transcript with the highest fold-change in C-33A RPTX cells was ABCB1, which encodes the MDR1 or P-gp protein. This increase in ABCB1 expression could be responsible for the reduction in the accumulation of the reporter drug doxorubicin and, at least in part, for the resistance to paclitaxel in C-33A RPTX cells. In turn, ABCB1 overexpression could be a consequence of the alteration of signaling pathways, such as ECM, MAPKs, and PI3K-Akt, in C-33A RPTX cells.
lncRNAs are capable of regulating gene expression by interacting with DNA, RNA, and proteins at multiple regulatory levels, including the regulation of the chromatin architecture and transcription, RNA processing, editing, localization, and stability; and protein translation and localization [55, 56]. In this work, KEGG analysis of lncRNAs also showed information about different signaling pathways that are associated with chemoresistance, such as adherens junctions, tight junctions, focal adhesion, apoptosis, MAPKs, EGFR tyrosine kinase inhibitor resistance, platinum drug resistance, and others. Other studies have accumulated evidence that lncRNAs have important roles in the sensitivity of tumor cells to chemotherapy, modifying the expression of key genes. For example, PCAT6 is a lncRNA known for its oncogenic activity in prostate cancer [57]. In a previous study, the overexpression of PCAT6 in cervical cancer accelerated proliferation, metastasis, and cisplatin resistance, with suppression of apoptosis through the PCAT6/miR-543/ZEB1 regulatory axis, with ZEB1 being a transcriptional regulator that promotes EMT [58]. In our C-33A RPTX cell model, the PCAT6 transcript was overexpressed, which could be driving the resistance phenotype through a ZEB1-independent pathway, since the latter was not differentially expressed. Another upregulated lncRNA in C-33A RPTX cells was TP53TG1, a controversial pro- and anti-tumoral regulator. In non- small lung cancer cells, the upregulation of TP53TG1 led to an enhanced cisplatin sensitivity and apoptosis of A549/DDP cells, while TP53TG1 depletion elicited the opposite effects, showing a sensitizing effect on these cells [59], which possibly depends on the cellular context, and therefore, in this model of resistance in cervical cancer cells, its overexpression represents a cause of resistance to paclitaxel. Meanwhile, a pan-cancer analysis showed that high expression of TP53TG1 was associated with a better prognosis in cervical cancer patients, and its overexpression in HeLa cervical cancer cells led to the inhibition of cell proliferation and promotion of apoptosis [60]. The opposite function of TP53TG1 was shown in hepatocellular carcinoma cells HepG2, in which siRNA- directed knockdown of the TP53TG1 sensitized them to the antiproliferative effects of Sorafenib. In addition, the combination of sorafenib and the downregulation of the TP53TG1 drastically reduced the phosphorylation of ERK MAPK [61]. In summary, these studies on lncRNAs demonstrate the plasticity of these types of biomolecules in regulating gene expression and their adaptation to the cellular context, possibly due to the multiple targets they can regulate at different levels and mechanisms through which they exert their function. Therefore, further in vitro and clinical studies are required to gain greater knowledge of their functions for the translational applications, such as the selection of lncRNAs as markers and pharmacological targets.
In Conclusion, the C-33A RPTX model offers an innovative experimental framework for investigating the molecular mechanisms underlying chemoresistance in cervical and possibly other types of cancer. This dual-level approach, from coding and long non-coding RNAs, provides a better perspective on paclitaxel resistance. Thus, this model is proposed as a biological platform for evaluating new antineoplastic agents and elucidating their mechanisms of action, thereby enhancing their translational relevance. Furthermore, the C-33A RPTX system enables the identification of potential biomarkers predictive of drug response, contributing to the development of personalized therapeutic strategies. Its comprehensive characterization also facilitates comparative studies with other chemoresistant phenotypes, offering insights into shared and unique adaptive molecular signatures. As a perspective, by integrating genomic, transcriptomic, and functional data, this model supports the exploration of regulatory networks that sustain drug tolerance and survival.
Acknowledgements
This work was supported by Autonomous University of Sinaloa for the financial support [PROFAPI PRO_ A2_014 and PROFAPI PRO_A2_015].
Conflict of interest
The authors declare no conflict of interest.
Authors’ contributions
AFC-S and MM-V conceived the original idea and planned the experiments of the project. AFC-S and NLD-T participated in the entire experiment. MC-H participated in the management of cell culture and the generation of chemoresistance. RB-R participated in the processing, acquisition, and analysis of the samples in the flow cytometry experiment. JRP-U and AG-G participated in the interpretation of bioinformatic analysis of the transcriptomics from the data provided by Novogene Corporation Inc. All authors analyzed and interpreted the data. OP-Z helped to supervise the project. AFC-S wrote the manuscript with the support of MM-V and OP-Z. All authors contributed to the final version of the manuscript.
References
- Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL , Soerjomataram I, Jemal A. CA: a cancer journal for clinicians.2024;74(3). CrossRef
- Battling Chemoresistance in Cancer: Root Causes and Strategies to Uproot Them Ramos A, Sadeghi S, Tabatabaeian H. International Journal of Molecular Sciences.2021;22(17). CrossRef
- Ion Channel Involvement in Tumor Drug Resistance Altamura C, Gavazzo P, Pusch M, Desaphy J. Journal of Personalized Medicine.2022;12(2). CrossRef
- Regulation of paclitaxel activity by microtubule-associated proteins in cancer chemotherapy Shi X, Sun X. Cancer Chemotherapy and Pharmacology.2017;80(5). CrossRef
- Relationship of mitotic arrest and apoptosis to antitumor effect of paclitaxel Milross C. G., Mason K. A., Hunter N. R., Chung W. K., Peters L. J., Milas L.. Journal of the National Cancer Institute.1996;88(18). CrossRef
- Nuclear Lamin A/C Expression Is a Key Determinant of Paclitaxel Sensitivity Smith ER , Leal J, Amaya C, Li B, Xu X. Molecular and Cellular Biology.2021;41(7). CrossRef
- Breaking malignant nuclei as a non-mitotic mechanism of taxol/paclitaxel Smith ER , Xu X. Journal of Cancer Biology.2021;2(4). CrossRef
- Paclitaxel directly binds to Bcl-2 and functionally mimics activity of Nur77 Ferlini C, Cicchillitti L, Raspaglio G, Bartollino S, Cimitan S, Bertucci C, Mozzetti S, et al . Cancer Research.2009;69(17). CrossRef
- Paclitaxel's Mechanistic and Clinical Effects on Breast Cancer Abu Samaan TM , Samec M, Liskova A, Kubatka P, Büsselberg D. Biomolecules.2019;9(12). CrossRef
- The lactate receptor (HCAR1/GPR81) contributes to doxorubicin chemoresistance via ABCB1 transporter up-regulation in human cervical cancer HeLa cells Wagner W., Kania K. D., Blauz A., Ciszewski W. M.. Journal of Physiology and Pharmacology: An Official Journal of the Polish Physiological Society.2017;68(4). CrossRef
- The ABCB1 3435C > T polymorphism influences docetaxel transportation in ovarian cancer Yin B, Lu P, Liang J, Zhang W, Xin M, Pei K, Li Y. The Journal of International Medical Research.2019;47(10). CrossRef
- Clinical significance and correlation of miR-200c and P-gp expression in gastric cancer and the effects on multidrug resistance Wang S, Guo J, Mo Z, Shi X, Qu C. Journal of Gastrointestinal Oncology.2022;13(2). CrossRef
- ABCB1 Regulates Immune Genes in Breast Cancer Chen H, Chen Y, Wang C, Chung W, Fang J, Lai M, Hsu H. Breast Cancer (Dove Medical Press).2023;15. CrossRef
- ABCB1 polymorphism predicts the toxicity and clinical outcome of lung cancer patients with taxane-based chemotherapy Zhong J, Guo Z, Fan L, Zhao X, Zhao B, Cao Z, Cheng L, et al . Thoracic Cancer.2019;10(11). CrossRef
- The inhibition of ABCB1/MDR1 or ABCG2/BCRP enables doxorubicin to eliminate liver cancer stem cells Yin W, Xiang D, Wang T, Zhang Y, Pham CV , Zhou S, Jiang G, et al . Scientific Reports.2021;11(1). CrossRef
- Increase of beta(III)- and beta(IVa)-tubulin isotopes in human prostate carcinoma cells as a result of estramustine resistance Ranganathan S., Dexter D. W., Benetatos C. A., Chapman A. E., Tew K. D., Hudes G. R.. Cancer Research.1996;56(11). CrossRef
- LncRNA H19 Regulates Breast Cancer DNA Damage Response and Sensitivity to PARP Inhibitors via Binding to ILF2 Zhao J, Xu J, Wu M, Wang W, Wang M, Yang L, Cai H, et al . International Journal of Molecular Sciences.2023;24(11). CrossRef
- Knockdown of lncRNA H19 restores chemo-sensitivity in paclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Akt signaling pathway Han J, Han B, Wu X, Hao J, Dong X, Shen Q, Pang H. Toxicology and Applied Pharmacology.2018;359. CrossRef
- Transcriptomic Landscape of Paclitaxel-Induced Multidrug Resistance in 3D Cultures of Colon Cancer Cell Line DLD1 Dragicevic S, Dinic J, Ugrin M, Vidovic M, Babic T, Nikolic A. International Journal of Molecular Sciences.2025;26(14). CrossRef
- Integrated transcriptome analysis and in silico investigations identify KHDRBS3 to target with steroidal lactone in paclitaxel resistance breast cancer Sarkar T, Sarkar T, Goswami M, Robaszkiewicz A, Sarkar K. Biochemical and Biophysical Research Communications.2025;777. CrossRef
- Cisplatin plus paclitaxel chemotherapy with or without bevacizumab in postmenopausal women with previously untreated advanced cervical cancer: a retrospective study Chu G, Liu X, Yu W, Chen M, Dong L. BMC cancer.2021;21(1). CrossRef
- Comparing Paclitaxel-Carboplatin with Paclitaxel-Cisplatin as the Front-Line Chemotherapy for Patients with FIGO IIIC Serous-Type Tubo-Ovarian Cancer Huang C, Cheng M, Lee N, Huang H, Lee W, Chang W, Wang P. International Journal of Environmental Research and Public Health.2020;17(7). CrossRef
- Carboplatin and paclitaxel as first-line treatment of unresectable or metastatic esophageal or gastric cancer Prithviraj G. K., Baksh K., Fulp W., Meredith K., Hoffe S., Shridhar R., Almhanna K.. Diseases of the Esophagus: Official Journal of the International Society for Diseases of the Esophagus.2015;28(8). CrossRef
- Overcoming Chemotherapy Resistance in Metastatic Cancer: A Comprehensive Review Eslami M, Memarsadeghi O, Davarpanah A, Arti A, Nayernia K, Behnam B. Biomedicines.2024;12(1). CrossRef
- Molecular mechanisms of cisplatin resistance in cervical cancer Zhu H, Luo H, Zhang W, Shen Z, Hu X, Zhu X. Drug Design, Development and Therapy.2016;10. CrossRef
- In vitro Development of Chemotherapy and Targeted Therapy Drug-Resistant Cancer Cell Lines: A Practical Guide with Case Studies McDermott M, Eustace AJ , Busschots S, Breen L, Crown J, Clynes M, O'Donovan N, Stordal B. Frontiers in Oncology.2014;4. CrossRef
- Omics Analysis of Chemoresistant Triple Negative Breast Cancer Cells Reveals Novel Metabolic Vulnerabilities Kordias D, Kostara CE , Papadaki St, Verigos J, Bairaktari E, Magklara A. Cells.2022;11(17). CrossRef
- Integrating transcriptomics, proteomics, glycomics and glycoproteomics to characterize paclitaxel resistance in breast cancer cells Cao L, Zhou Y, Li X, Lin S, Tan Z, Guan F. Journal of Proteomics.2021;243. CrossRef
- Ginsenoside Rg5 Sensitizes Paclitaxel-Resistant Human Cervical-Adeno-Carcinoma Cells to Paclitaxel-And Enhances the Anticancer Effect of Paclitaxel Ramesh J, Thilakan RC , Gopalakrishnan RM , Vijayapoopathi S, Dorschel A, Venugopal B. Genes.2022;13(7). CrossRef
- CENPU affects the paclitaxel resistance of cervical cancer cells by regulating the FOXM1/ABCC5 pathway Zhang X, Zhang Y, Wang M. European Journal of Gynaecological Oncology.2024;45(1):89-95.
- CD133+/ABCC5+ cervical cancer cells exhibit cancer stem cell properties He L, Qian H, Seyiti A, Yang C, Shi N, Chen C, Zhang P, Hou Y. Heliyon.2024;10(17). CrossRef
- Neuronal calcium signaling: function and dysfunction Brini M, Calì T, Ottolini D, Carafoli E. Cellular and molecular life sciences: CMLS.2014;71(15). CrossRef
- Targeting Intracellular Calcium Signaling ([Ca2+]i) to Overcome Acquired Multidrug Resistance of Cancer Cells: A Mini-Overview Büsselberg D, Florea A. Cancers.2017;9(5). CrossRef
- Calcium Signaling Regulates Autophagy and Apoptosis Sukumaran P, Nascimento Da Conceicao V, Sun Y, Ahamad N, Saraiva LR , Selvaraj S, Singh BB . Cells.2021;10(8). CrossRef
- Oncogenes in cell survival and cell death Shortt J, Johnstone RW . Cold Spring Harbor Perspectives in Biology.2012;4(12). CrossRef
- Endoplasmic Reticulum-Mitochondrial Ca2+ Fluxes Underlying Cancer Cell Survival Ivanova H, Kerkhofs M, La Rovere RM , Bultynck G. Frontiers in Oncology.2017;7. CrossRef
- Paclitaxel affects cytosolic calcium signals by opening the mitochondrial permeability transition pore Kidd JF , Pilkington MF , Schell MJ , Fogarty KE , Skepper JN , Taylor CW , Thorn P. The Journal of Biological Chemistry.2002;277(8). CrossRef
- Emerging molecular mechanisms in chemotherapy: Ca2+ signaling at the mitochondria-associated endoplasmic reticulum membranes Kerkhofs M, Bittremieux M, Morciano G, Giorgi C, Pinton P, Parys JB , Bultynck G. Cell Death & Disease.2018;9(3). CrossRef
- ERK/MAPK signalling pathway and tumorigenesis Guo Y, Pan W, Liu S, Shen Z, Xu Y, Hu L. Experimental and Therapeutic Medicine.2020;19(3). CrossRef
- Targeting the RAS/RAF/MAPK pathway for cancer therapy: from mechanism to clinical studies Bahar ME , Kim HJ , Kim DR . Signal Transduction and Targeted Therapy.2023;8(1). CrossRef
- FGF2 is overexpressed in asthma and promotes airway inflammation through the FGFR/MAPK/NF-κB pathway in airway epithelial cells Tan Y, Zhou H, Lin Y, Yi L, Chen Z, Cao Q, Guo Y, et al . Military Medical Research.2022;9(1). CrossRef
- Tephrosin Suppresses the Chemoresistance of Paclitaxel-Resistant Ovarian Cancer via Inhibition of FGFR1 Signaling Pathway Kim HS , Bae S, Lim YJ , So KA , Kim TJ , Bae S, Lee JH . Biomedicines.2023;11(12). CrossRef
- The p38/MAPK pathway regulates microtubule polymerization through phosphorylation of MAP4 and Op18 in hypoxic cells Hu J, Chu Z, Han J, Dang Y, Yan H, Zhang Q, Liang G, Huang Y. Cellular and molecular life sciences: CMLS.2010;67(2). CrossRef
- Inhibition of p38-MK2 pathway enhances the efficacy of microtubule inhibitors in breast cancer cells Chen Y, Takada M, Nagornyuk A, Yu M, Yamada H, Nagashima T, Ohtsuka M, et al . eLife.2025;13. CrossRef
- The Rap1-RIAM-talin axis of integrin activation and blood cell function Lagarrigue F, Kim C, Ginsberg MH . Blood.2016;128(4). CrossRef
- The Role of Ras-Associated Protein 1 (Rap1) in Cancer: Bad Actor or Good Player? Looi C, Hii L, Ngai S, Leong C, Mai C. Biomedicines.2020;8(9). CrossRef
- Rap1 coordinates cell-cell adhesion and cytoskeletal reorganization to drive collective cell migration in vivo Rothenberg KE , Chen Y, McDonald JA , Fernandez-Gonzalez R. Current biology: CB.2023;33(13). CrossRef
- Circ_0000231 promotes paclitaxel resistance in ovarian cancer by regulating miR-140/RAP1B Liu J, Wang H, Xiao S, Zhang S, Qi Y, Wang M. American Journal of Cancer Research.2023;13(3). CrossRef
- Polarity proteins in migration and invasion Etienne-Manneville S.. Oncogene.2008;27(55). CrossRef
- Prostaglandins in cancer cell adhesion, migration, and invasion Menter DG , Dubois RN . International Journal of Cell Biology.2012;2012. CrossRef
- The Interplay between Extracellular Matrix Remodeling and Cancer Therapeutics Prakash J, Shaked Y. Cancer Discovery.2024;14(8). CrossRef
- The extracellular matrix alteration, implication in modulation of drug resistance mechanism: friends or foes? Jurj A, Ionescu C, Berindan-Neagoe I, Braicu C. Journal of experimental & clinical cancer research: CR.2022;41(1). CrossRef
- Extracellular Matrix Proteins Confer Cell Adhesion-Mediated Drug Resistance Through Integrin α v in Glioblastoma Cells Yu Q, Xiao W, Sun S, Sohrabi A, Liang J, Seidlits SK . Frontiers in Cell and Developmental Biology.2021;9. CrossRef
- β1-Integrin binding to collagen type 1 transmits breast cancer cells into chemoresistance by activating ABC efflux transporters Baltes F, Pfeifer V, Silbermann K, Caspers J, Wantoch von Rekowski K, Schlesinger M, Bendas G. Biochimica Et Biophysica Acta. Molecular Cell Research.2020;1867(5). CrossRef
- Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, Shen F. International Journal of Molecular Sciences.2019;20(22). CrossRef
- Gene regulation by long non-coding RNAs and its biological functions Statello L, Guo C, Chen L, Huarte M. Nature Reviews. Molecular Cell Biology.2021;22(2). CrossRef
- The Role of lncRNA PCAT6 in Cancers Wang S, Chen Z, Gu J, Chen X, Wang Z. Frontiers in Oncology.2021;11. CrossRef
- LncRNA PCAT6 Accelerates the Progression and Chemoresistance of Cervical Cancer Through Up-Regulating ZEB1 by Sponging miR-543 Ma Z, Gu G, Pan W, Chen X. OncoTargets and Therapy.2020;13. CrossRef
- TP53TG1 enhances cisplatin sensitivity of non-small cell lung cancer cells through regulating miR-18a/PTEN axis Xiao H, Liu Y, Liang P, Wang B, Tan H, Zhang Y, Gao X, Gao J. Cell & Bioscience.2018;8. CrossRef
- LncRNA TP53TG1 plays an anti-oncogenic role in cervical cancer by synthetically regulating transcriptome profile in HeLa cells Cheng Y, Huang N, Yin Q, Cheng C, Chen D, Gong C, Xiong H, et al . Frontiers in Genetics.2022;13. CrossRef
- Knockdown of lncRNA TP53TG1 Enhances the Efficacy of Sorafenib in Human Hepatocellular Carcinoma Cells Lu Q, Xin M, Guo Q, Rothberg BS , Gamero AM , Yang L. Non-coding RNA.2022;8(4). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2026
Author Details