Poly (ADP-ribose) Polymerase Promoter Hypermethylation Predispose Females to Breast Cancer
Download
Abstract
Being the most common cause of female deaths worldwide, breast cancer (BC) is intensively studied over the last two decades. In the present investigation, we evaluated the promoter methylation of three cancer-related genes; PARP-1, p21, and Rb in 10 bi-matched BC samples (ductal carcinoma and lobular carcinoma) included the core tumor and the adjacent normal tissue. H&E-stained histopathological sectioning revealed grade 2 and grade 3 tumor cells. Methylation-specific PCR (MSP) was performed using methylated (M) and unmethylated (U) primers for the three genes understudy. Histone acetyltransferase was measured in tumor and healthy tissues. A variation in the methylation state of the promoter region of the three genes were observed in core tumor and healthy tissue. PARP and Rb were hypermethylated in tumor tissues while p21 was partially methylated. HAT activities were positively correlated with the methylation pattern observed in healthy tissues, as HAT was highly expressed in healthy vs. tumor tissues. The obtained data might indicate that patients might be at risk of BC recurrence after being subjected to mastectomy. These data could be employed as a core in epigenetic-based data mining to establish a model for predicting the breast cancer-predisposed patients. However, further investigations are needed to fulfill this goal.
Introduction
Breast cancer (BC) is considered one of the most prevailed cancer in women worldwide, with very high mortality rate [1] . There is persistent increase in the global incidence rate of breast cancer, which call for urgent global management, particularly in middle-income countries [2] . Epigenetic modifications i.e., DNA methylation and chromatin remodeling have proven to play a pivotal role since the early stages of breast cancerogenesis [3] .
PARPs are a family that comprises 18-membered of proteins function to post-translationally modify the target proteins [4] . PARP-1 is the most important and extensively studied member of this family (which include PARP1, PARP2, PARP3, PARP4, PARP-5a, PARP-5b, PARP6, PARP7, PARP8, PARP9, PARP10, PARP11, PARP12, PARP14, PARP15, and PARP16), where it is responsible for the majority of poly (ADP-ribosyl)ation processes in the cells [5] . PARPs are responsible for the early response of mammalian cells to the DNA damage caused by either endogenous or exogenous agents. PARP is being activated upon single or double strand DNA breaks, and it is upregulated when cells treated with any DNA-damaging agents [6] . Several environmental agents have proven to cause severe downregulation of PARP expression with an increased level of methylation in the promoter region; these agents include benzene [7] and nano-SiO2 [6] .
The cyclin-dependent kinase inhibitor (CDKN), p21, is well known for its crucial role in enforcing cell cycle to arrest [8] , and as a negative regulator of the G1/S transition. It has an inhibitory effect on a wide range of cyclin-CDKs [9] . Despite its role in ceasing cellular proliferation, many studies suggested that p21 might, in the same time, enhance cellular proliferation, senescence, oncogenicity and it might have an anti-apoptotic function [10] .
Retinoblastoma (Rb) is regulator of the division of cancer cells, and it is involved in many diseases as well as in remodeling of chromatin [11] . It also controls tumor progression, as it oversees a G1 checkpoint that stops S-stage entry and further cellular growth [12] . The loss of Rb functionality induces cancer initiation, as the cell cycle being deregulated [3] . In human cancers (e.g. retinoblastoma, osteosarcoma, and lung carcinoma), Rb is downregulated either by direct mutations such as deletion, inversion or by hypermethylation of its promoter region [14] .
Epigenetic changes are the most common molecular changes in human cancer. Aberrant promoter methylation in cytosine-phosphate-guanine (CpG) islands takes place in several genes in cancer development and progression [15] . It was suggested that these CpG island methylation of tumor-related genes is considered a very early molecular event in the progression of breast cancer [16].
Investigating epigenetic mechanisms of the regulation of gene expression offers a novel approach for diagnosis and treatment of cancer [17] . It has been indicated that promoter hypermethylation has a strong inverse correlation with gene expression with estimates varying from 600 to 1000 hypo- or hypermethylated genes per tumor [18] [19] . Hypermethylation is, for that reason, considered the hallmark of human cancer [20] .The present study aimed to assess the level of methylation of PARP-1, p21, and Rb gene promoters in breast cancer female patients.
Methods
Tissue samples
Ten bi-matched BC (core tumor and safety margin) tissues were collected from patients subjected mastectomy at Alaa Ezzat Hospital, Cairo, Egypt. We obtained written informed consents from all participants indicating that their removed tissues will be used in research purposes only. About 0.5 gram of tumor along with 0.5 gram of healthy tissues (safety margin) was collected from each participant. Samples were kept directly in PBS until being used in DNA and nuclear protein extractions. Participants with family history and those who smoke heavily were excluded from this study.
Histopathological sections
For each tissue, histopathological sections were made using ultra-microtome sectioning machine. The slides were prepared using the standard procedures in which tissues were fixed with formalin fixation and embedded on paraffin, and then cut in a thickness of 3-5 µm sections and staining with H & E. Sections were fixed on glass slides and analyzed under light microscope.
Tumor grading
Post-operative tumor grading was performed. The grading system used was Three-tier grading scheme.
DNA extraction
Genomic DNA was extracted from tissue samples using QIAamp DNA Mini Kit (Qiagen, Germany). The procedures were performed according to the manufacturer’s protocol.
Nuclear protein extraction
Nuclear protein was extracted using a manual method. The procedures were as follows: 0.5 g of tissue was grinded in pre-chilled mortar and in the presence of 1.5 mL of PBS as sample buffer and autoclaved sand. Lysates were transferred to Eppendorf tubes and centrifuged for 4 min at 2000 rpm to precipitate cell debris and sand. The supernatant (contains the nuclear protein) was collected and suspended in 62 µL BL (Tris pH 8.0, LiCl, and glycerol) buffer and incubated on ice for 5 minutes. The nuclear proteins were then ready to further analysis.
DNA bisulfite conversion
Extracted DNA was converted using EZ DNA Methylation-LightningTM (Zymo Research, USA). The procedures were performed according to the kit’s protocol.
Methylation-specific PCR (MSP)
After converting DNA, samples were subjected to specific PCR against six sets of primers representing three different genes. Each primer was designed in two instead of one set to detect both the methylated and unmethylated promoter region of the genes under study. The amplifications were consisted of a denaturation (95°C for 5 min) and then 35 cycles (95°C for 30s, 58±5—depends on the gene—for 45s and 72°C for 30s). A final extension step was performed at 72°C for 10 min. PCR mixture consisted of 100-200 ng of converted DNA, 1 x Master Mix (Cinnagen Inc., Tehran, Iran), 10 pmol of forward and reverse primers. The final volume was brought to 25 µL using dH2O. Amplified products were electrophoresed on 2% agarose gel and visualized by ethidium bromide staining. All MSP assays were repeated at least twice. The primers used in this study were PARP, P21, and Rb, and the sequences are listed in Tabl. 1.
| Sequence (5′ to 3′) | Primer |
| TTGTGGACGGTAGGTTAGAAC | PARP-1-M-F |
| AACTAAATCCGAAAAACGCA | PARP-1-M-R |
| GGTTTGTGGATGGTAGGTTAGAAT | PARP-1-U-F |
| AACAACTAAATCCAAAAAACACA | PARP-1-U-R |
| TACGCGAGGTTTCGGGATC | P21-M-F |
| CCCTAATATACAACCGCCCCG | P21-M-R |
| GGATTGGTTGGTTTGTTGGAATTT | P21-U-F |
| ACAACCCTAATATACAACCACCCCA | P21-U-R |
| GGTTTCGTTTTTTATGGTCGGGTACGGTTTACG | Rb-M-F |
| AAAAACGTAAAAACGACGACCATACC | Rb-M-R |
| GGTTTTGTTTTTTATGGTTGGGTATGGTTTATG | Rb-U-F |
| CATAAAAACAACAACCATACC | Rb-U-R |
Measuring the HAT activity
Total histone acetyl transferase (HAT) activity/inhibition was measured using the EpiQuik™ kit (Epigentek, USA). We followed the kit’s protocol.
Statistical analysis
Statistical analyses were performed using SPSS version 16 software package (SPSS Inc, Chicago, IL). Associations between methylation of loci, clinical and biological features were evaluated with chi-square and Fishers exact test.
Results
Histopathological perspectives
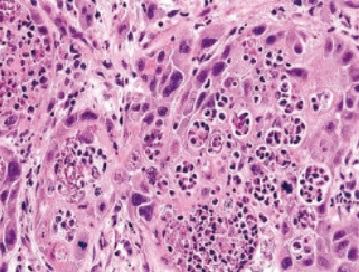
In the present study, all sections of the tumors were stained with H&E and examined under light microscope. Histopathological sectioning of the tumor samples revealed a G-II and G-III tumors using the three-tier scheme. The core samples were of grade 2 and 3 (Fig. 1 and 2).
Figure 1 :G-II H&E stained section showing solid groups of large atypical ductal cells exhibiting moderate anaplasia and mitosis. Focal ductal differentiation is seen. X100.
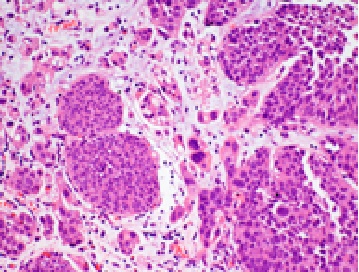
Figure 2 :G-III H&E stained section showing solid groups of large atypical ductal cells exhibiting moderate anaplasia and mitosis. X400.
Methylation Specific PCR
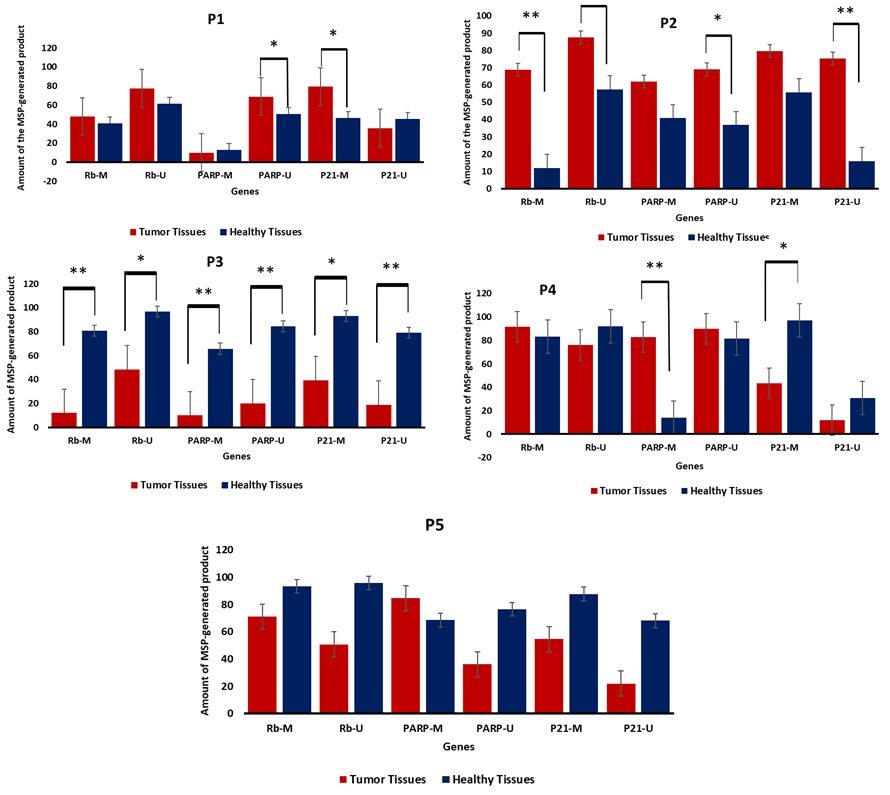
Methylation Specific PCR was performed to identify the methylation patterns related to the malignant and non-malignant tissues. Rb and PARP , and a slightly normal methylation pattern of p21. For Rb and PARP , the promoter regions were hypermethylated in almost 60% of the cases studied, while p21 promoter methylation was noticed in 40% of the cases. Meanwhile, a methylation pattern was also reported in healthy tissue (taken from safety margins) and this might serve as a marker for the future transformation of this tissue into malignant state. Methylation profile of cases was positively correlated with histopathological perspectives and tumor stages (data not shown). Ten samples were obtained from five breast cancer female patients (10 b-matched samples) and were all subjected to MSP. The results obtained indicated a variable methylation pattern of each of the three genes understudy (Fig. 3 and 4). Our results showed a of hypermethylation profile in Rb and PARP, and a slightly normal methylation pattern of p21. For Rb and PARP, the promoter regions were hypermethylated in almost 60% of the cases studied, while p21 promoter methylation was noticed in 40% of the cases. Meanwhile, a methylation pattern was also reported in healthy tissue (taken from safety margins) and this might serve as a marker for the future transformation of this tissue into malignant state. Methylation profile of cases was positively correlated with histopathological perspectives and tumor stages (data not shown).
Figure 3 :The MSP results showed the generated bands after being electrophoresed on 2% agarose gel.P1-P5: patient numbers. M: methylated primer, U: unmethylated primer.
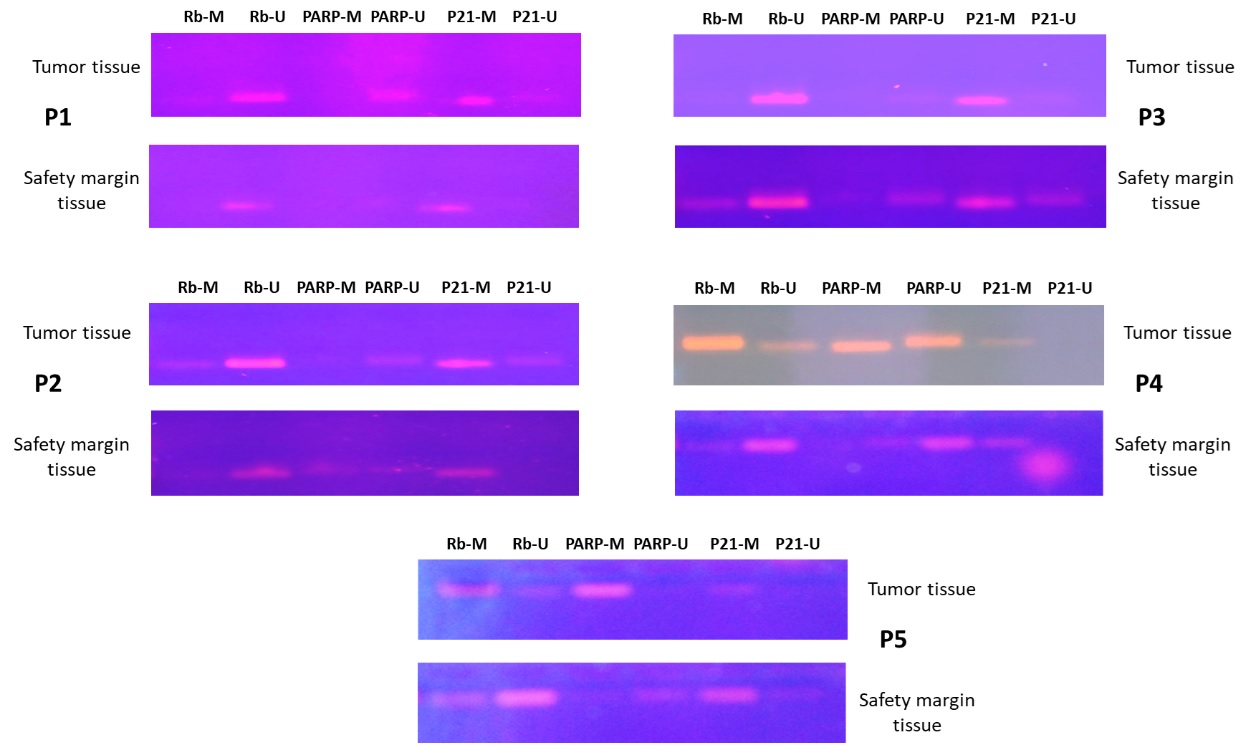
Figure 4 :The amount of the MSP-generated bands from breast cancer/healthy tissues against three tumor suppressor genes; Rb , PARP , and p21 .
HAT activity
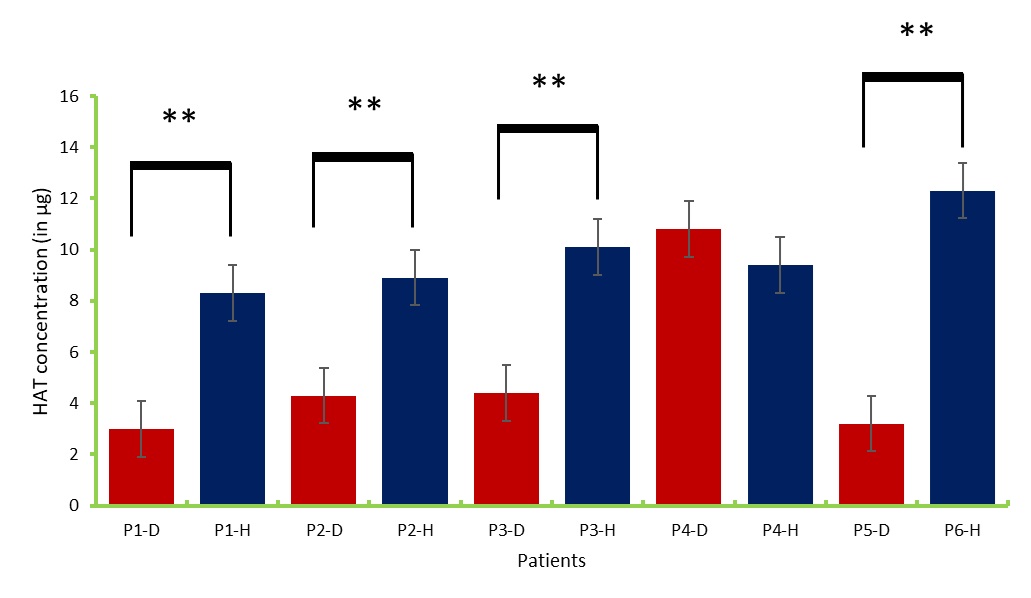
Nuclear protein was extracted and subjected to ELISA-like assay to measure the activity of histone acetyltransferases (HAT) in both tumor and safety margin tissues (Fig 5and 6). Data obtained indicated that histone acetyltransferases concentration was higher in the safety margin tissues compared to the core tumor samples. This might indicate the role of HAT is opening the chromatin structure in the genes in this study. HAT levels had also negative correlation with the methylation pattern; the high the methylation levels the low the HAT activity.
Figure 5 :Histone acetyltransferase standard curve
Figure 6 :The concentration of histone acetyltransferase in the core tumor and healthy tissues. D: diseased, H: healthy.
Discussion
Methylation Specific PCR
The present study investigated the methylation patterns of three tumor suppressor genes; PARP, Rb, and p21 in breast core tumor and healthy 10 bi-matched samples. Methylation specific PCR is a reliable method to detect the pattern of methylation of any gene. Here we detected a variable methylation profile in the three genes under study. It was indicated that Rb [21] and PARP [22] were hypermethylated in breast cancer patients. Both Rb and PARP were hypermethylated in 60% of the cases studied, while p21 promoter methylation was hypermethylated in the remaining 40%. This pattern was indicated previously [23, 24] . Meanwhile, a slight methylation score was also reported in normal tissues taken from the safety margin around the core tumor. This could be considered as prognosis for the disease or as a genetic marker for the future transformation of normal tissues into malignant [25] . Many research groups [25, 26] have indicated obtaining TSGs promoter methylation in safety margin tissues. This study, along with others, might help generating an epigenetic-based model to predict breast cancer in predisposed women.
HAT activity
Histone acetyltransferase modifies histones and is considered a major player in the epigenetic modulation of gene transcription [27, 28]. HATs are well known for their function to open chromatin structure and hence, facilitate the transcription of the gene where it resides [29] . In the present study, HAT was found to be highly expressed in all normal samples, which might indicate that the tumor suppressor genes under study along with apoptosis-related genes were functional and that might have led to the normal state of the breast cells [8,30]. These data support the notion of using HAT activator as a chemotherapeutic option in treating breast cancer [30, 31]. However, these data urge further studies to correlate the epigenetic readings with the tumor stage/grade in a model to predict the BC in predisposed women.
References
[1]. Lopes-Coelho F, Andre S, Felix A, Serpa J. Breast cancer metabolic cross-talk: Fibroblasts are hubs and breast cancer cells are gatherers of lipids. Molecular and cellular endocrinology. 2018;462(Pt B):93-106.
[2]. Carioli G, Malvezzi M, Rodriguez T, Bertuccio P, Negri E, La Vecchia C. Trends and predictions to 2020 in breast cancer mortality: Americas and Australasia. Breast (Edinburgh, Scotland). 2018;37:163-9.
[3]. Nowsheen S, Aziz K, Tran PT, Gorgoulis VG, Yang ES, Georgakilas AG. Epigenetic inactivation of DNA repair in breast cancer. Cancer letters. 2014;342(2):213-22.
[4]. Bai P, Canto C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell metabolism. 2012;16(3):290-5.
[5]. Dharwal V, Naura AS. PARP-1 inhibition ameliorates elastase induced lung inflammation and emphysema in mice. Biochemical pharmacology. 2018;150:24-34.
[6]. Gong C, Tao G, Yang L, Liu J, Liu Q, Li W, et al. Methylation of PARP-1 promoter involved in the regulation of nano-SiO2-induced decrease of PARP-1 mRNA expression. Toxicology letters. 2012;209(3):264-9.
[7]. Gao A, Zuo X, Liu Q, Lu X, Guo W, Tian L. Methylation of PARP-1 promoter involved in the regulation of benzene-induced decrease of PARP-1 mRNA expression. Toxicology letters. 2010;195(2-3):114-8.
[8]. Moussa RS, Kovacevic Z, Bae DH, Lane DJR, Richardson DR. Transcriptional regulation of the cyclin-dependent kinase inhibitor, p21(CIP1/WAF1), by the chelator, Dp44mT. Biochimica et biophysica acta General subjects. 2018;1862(3):761-74.
[9]. Nishiyama H, Gill JH, Pitt E, Kennedy W, Knowles MA. Negative regulation of G(1)/S transition by the candidate bladder tumour suppressor gene DBCCR1. Oncogene. 2001;20(23):2956-64.
[10]. Taniguchi K, Matsumura K, Kageyama S, Ii H, Ashihara E, Chano T, et al. Prohibitin-2 is a novel regulator of p21(WAF1/CIP1) induced by depletion of gamma-glutamylcyclotransferase. Biochemical and biophysical research communications. 2018;496(1):218-24.
[11]. Murphree AL, Benedict WF. Retinoblastoma: clues to human oncogenesis. Science. 1984;223(4640):1028-33.
[12]. Duggal S, Jailkhani N, Midha MK, Rao KVS, Kumar A. Rb interactome data and its modulations during cell cycle progression in HEK 293 cells. Data in brief. 2018;17:604-9.
[13]. Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25(38):5220
[14]. Liu H, Dibling B, Spike B, Dirlam A, Macleod K. New roles for the RB tumor suppressor protein. Current opinion in genetics & development. 2004;14(1):55-64.
[15]. Kim C, Paik S. Gene-expression-based prognostic assays for breast cancer. Nature reviews Clinical oncology. 2010;7(6):340-7.
[16]. Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW, Kim JH, et al. Promoter CpG island hypermethylation during breast cancer progression. Virchows Archiv : an international journal of pathology. 2011;458(1):73-84.
[17]. Cock-Rada A, Weitzman JB. The methylation landscape of tumour metastasis. Biology of the cell. 2013;105(2):73-90.
[18]. Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315-22.
[19]. Ushijima T, Asada K. Aberrant DNA methylation in contrast with mutations. Cancer science. 2010;101(2):300-5.
[20]. Portela A, Liz J, Nogales V, Setien F, Villanueva A, Esteller M. DNA methylation determines nucleosome occupancy in the 5'-CpG islands of tumor suppressor genes. Oncogene. 2013;32(47):5421-8.
[21]. Hajiloo M, Damavandi B, Hooshsadat M, Sangi F, Mackey JR, Cass CE, et al. Breast cancer prediction using genome wide single nucleotide polymorphism data. BMC bioinformatics. 2013;14 Suppl 13:S3.
[22]. Garber K. PARP inhibitors bounce back. Nature reviews Drug discovery. 2013;12(10):725.
[23]. Coppede F, Tannorella P, Stoccoro A, Chico L, Siciliano G, Bonuccelli U, et al. Methylation analysis of DNA repair genes in Alzheimer's disease. Mechanisms of ageing and development. 2017;161(Pt A):105-11.
[24]. Witkiewicz AK, Chung S, Brough R, Vail P, Franco J, Lord CJ, et al. Targeting the Vulnerability of RB Tumor Suppressor Loss in Triple-Negative Breast Cancer. Cell reports. 2018;22(5):1185-99.
[25]. Chi HC, Tsai CY, Tsai MM, Lin KH. Impact of DNA and RNA Methylation on Radiobiology and Cancer Progression. International journal of molecular sciences. 2018;19(2).
[26]. Sandoval-Basilio J, Gonzalez-Gonzalez R, Bologna-Molina R, Isiordia-Espinoza M, Leija-Montoya G, Alcaraz-Estrada SL, et al. Epigenetic mechanisms in odontogenic tumors: A literature review. Archives of oral biology. 2018;87:211-7.
[27]. Lu R, Wang GG. Tudor: a versatile family of histone methylation 'readers'. Trends in biochemical sciences. 2013;38(11):546-55.
[28]. Friedmann DR, Marmorstein R. Structure and mechanism of non-histone protein acetyltransferase enzymes. The FEBS journal. 2013;280(22):5570-81.
[29]. McGrath J, Trojer P. Targeting histone lysine methylation in cancer. Pharmacology & therapeutics. 2015;150:1-22.
[30]. You Y, Sawikowska A, Neumann M, Pose D, Capovilla G, Langenecker T, et al. Temporal dynamics of gene expression and histone marks at the Arabidopsis shoot meristem during flowering. Nature communications. 2017;8:15120.
[31]. Perez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics. 2017;12(5):323-39.
License
Copyright
© ,
Author Details