Effect of Trichostatin A on Histone Deacetylases 1, 2 and 3, p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2 Gene Expression in Breast Cancer SK-BR-3 Cell Line
Download
Abstract
Objective: DNA methylation, the covalent addition of a methyl group to cytosine, and histone modification play an important role in the establishment and maintenance of the program of gene expression. The balance of histone acetylation is determined by the activities of two groups of enzymes including histone acetyltransferases (HATs) and histone deacetylases (HDACs). Histone deacetylation is generally associated with silencing gene expression resulting in several solid tumors. HDAC inhibitors (HDACIs) are the new class of potential anticancer compounds for the treatment of the solid and hematological cancers. The current study was designed to evaluate the effect of trichostatin A (TSA) on histone deacetylases 1, 2 and 3, p21Cip1/Waf1/Sdi1 (p21), p27Kip1 (p27), and p57Kip2 (p57) gene expression in breast cancer SK-BR-3 cell line.
Materials and Methods: The breast cancer SK-BR-3 line was treated with TSA. To determine cell viability, cell apoptosis, and the relative expression level of the genes, MTT assay, cell apoptosis assay, and qRT-PCR were done respectively.
Results: TSA significantly inhibited cell growth, and induced apoptosis. Furthermore, this compound increased p21, p27, and p57 and decreased histone deacetylases 1, 2 and 3 gene expression significantly.
Conclusion: The TSA can reactivate the p21, p27, and p57 through down-regulation of histone deacetylases 1, 2 and 3 gene expression.
Introduction
DNA methylation, the covalent addition of a methyl group to cytosine, and histone modification play an important role in the establishment and maintenance of the program of gene expression. Hypermethylation of tumor suppressor genes (TSGs) are well documented in various cancers [1].The balance of histone acetylation and deactylation is determined by the activities of two groups of enzymes including histone acetyltransferases (HATs) and histone deacetylases (HDACs). Histone deacetylation are generally associated with silencing gene expression resulting in several solid tumours and haematological malignancies [2]. HDACs include a family of 18 genes, which are grouped into classes I–IV based on their homology to their respective yeast orthologues. Classes I, II, and IV consist of 11 family members, which are referred to as “classical” HDACs, whereas the 7 class III members are called sirtuins [3].
Overexpression of HDAC1, 2 and 3 has been reported in breast cancer [4]. In addition to breast cancer, upregulation of HDACs has been reported in other cancers. Several studies have indicated that class I HDAC isoforms 1 and 2 are highly expressed in renal and urothelial carcinomas [5-6]. Semilar mechanism has been shown in prostate cancer [7]. Mammillary Cell cycle progression is a regulated process that involves several checkpoints that assess extracellular signals and DNA integrity. Cyclins and their partner, cyclin-dependent kinases (CDKs), are positive regulators of cell cycle progression; whereas CDK inhibitors (CDKIs) are important negative regulators. The abnormal expression of CDKs or CDKIs leads to tumorigenesis. There are two types of CDKIs, including the CIP/KIP family, p21Cip1/Waf1/Sdi1 (p21), p27Kip1 (p27), and p57Kip2 (p57), and the INK family, p16, p15, p18, and p19. HDACs activity can affect the expression of CDKIs by deacetylation of these genes resulting in genes silenced and cancer induction [8].
HDAC inhibitors (HDACIs) are the new class of potential anticancer compounds for the treatment of solid and hematological cancers. These agents vary in structure and exert various anticancer activities such as cell cycle arrest and cell apoptosis. They are classified into hydroxamic acids (such as trichostatin A [TSA]), short-chain fatty acids (such as valproic acid [VPA] and phenylbutyrate), cyclic peptides (such as depsipeptide), and synthetic benzamides (such as MS-275). HDACI TSA was one of the first natural compounds determined to inhibit HDACs [9]. It has been shown that TSA induces apoptosis in colon cancer HCT116 cells [10]. Previously, we reported that TSA and VPA, as HDACIs, can induce apoptosis in HCC and colon cancer, respectively [11-12]. The current study was designed to evaluate the effect of TSA on histone deacetylases 1, 2, and 3, p21, p27, and p57 gene expression in breast cancer SK-BR-3 cell line.
Materials and Methods
Materials
The human breast cancer SK-BR-3 cell line was obtained from the National Cell Bank of Iran-Pasteur Institute and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with antibiotics and fetal bovine serum 10% in a humidified atmosphere of 5% CO2 in air at 37℃. All materials and compounds including TSA, various necesary kits, and instruments used in cell culture, flow cytometry assay, 3 (4,5 dimethyl 2 thiazolyl) 2, 5 diphenyl 2H tetrazolium bromide (MTT) assay, RNA extraction, reverse transcription, and real-time PCR were provided as obtained previously [12-13-14].
Cell culture and cell viability
The breast cancer SK-BR-3 cell viability was determined using MTT assay. The cells were cultured and seeded in 96-well plates at a density of 3 × 105 cells per well and treated with TSA (0.5, 1, 5, and 10 μM) dissolved in DMSO for 24 and 48 h. The control groups received the solvent of the drug, DMSO. After incubation for different periods, the cells were exposed to MTT, MTT (0.5 mg/mL) was added to each well for 4 h. Finally, the MTT metabolite was dissolved in DMSO (200 mL) and the optical density was detected by a microplate reader at a wavelength of 570 nM. Each experiment was repeated three times (triplicates).
Cell apoptosis assay
To determine the apoptotic breast cancer SK-BR-3 cells, the cells were cultured and seeded at a density of 3 × 105 cells/well and treated with TSA (1 μM) for 24 and 48 h. Following treatment, all treated and untreated cells were harvested by trypsin and washed twice with PBS and then resuspended in Binding buffer (1x). Finally, the cells were stained with annexin V-FITC (5 μl) and propidium iodide (5 μl) in the dark at room temperature for 15 min and counted by FACScan flow cytometry (Becton Dickinson, Heidelberg, Germany).
Real-time Quantitative Reverse Transcription Polymerase
Chain Reaction (qRT-PCR)
To determine the relative expression level of histone deacetylases 1, 2, and 3, p21, p27, and p57 gene expression genes expression, the qRT-PCR was performed. The SK-BR-3 cells were cultured at a density of 3 × 105 cells/well and treated with TSA (1 μM) for 24 and 48 h, based on IC50 values. After 24 and 48 h of treatment, the total RNA was harvested using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer protocol and treated by RNase-free DNase (Qiagen). The RNA was transcribed to complementary DNA (cDNA). QRT-PCR was done as described previously [15]. The program for the PCR was as we reported previously [16]. The primer sequences of the genes are indicated in Table1.
| Primer name | Primer sequences (5' to 3') | References |
| P21 Forward | AGG CGC CAT GTC AGA ACC GGC TGG | [17] |
| P21 Reverse | GGA AGG TAG AGC TTG GGC AGG C | [17] |
| P 27 Forward | ATG TCA AAC GTG CGA GTG TCT AAC | [17] |
| P 27 Reverse | TTA CGT TTG ACG TCT TCT GAG GCC A | [17] |
| P 57 Forward | GCGGCGATCAAGAAGCTGTC | [17] |
| P 57 Reverse | CCGGTTGCTGCTACATGAAC | [17] |
| GAPDH Forward | TCCCATCACCATCTTCCA | [18] |
| GAPDH Reverse | CATCACGCCACAGTTTCC | [18] |
| HDAC1 Forward | GACACGCCAAGTGTGTGGAA | [19] |
| HDAC1 Reverse | CCTCCCAGCATCAGCATAGG | [19] |
| HDAC2 Forward | ACATGAGCAATGCGGAGAAAT | [19] |
| HDAC2 Reverse | TCTGCCATCTTGTGGTACAGTGA | [19] |
| HDAC3 Forward | CCTTTTCCAGCCGGTTATCA | [19] |
| HDAC3 Reverse | ACAATGCACGTGGGTTGGT | [19] |
GAPDH was used as an endogenous control. Data were analyzed using the comparative Ct (ΔΔct) method.
Statistical analysis
The database was setup with the SPSS 16.0 software package (SPSS Inc., Chicago, Illinois, USA) for analysis. The data were acquired from three tests and are shown as means ± standard deviations. Statistical comparisons between groups were performed with ANOVA (one-way ANOVA) and Turkey test. A significant difference was considered as P < 0.05.
Results
Result of cell viability by the MTT assay
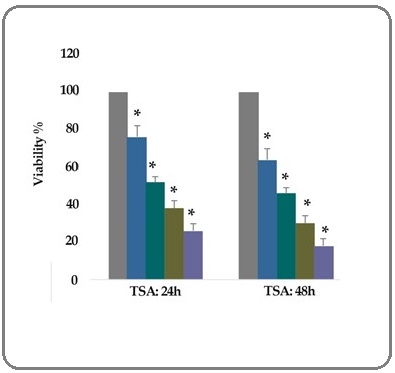
To evaluate the effect of TSA (0.5, 1, 5, and 10 μM) on the breast cancer SK-BR-3 cell viability, MTT assay was utilized. Our results indicated that the rate of cell growth inhibition was significantly increased in treated groups than that in control groups after 24 and 48 h. Results showed that the number of viable SK-BR-3 cells decreased significantly, as the concentration of the compounds and duration increased; indicating a dose- and duration-dependent relationship (P< 0.001), as shown in Figure 1. The IC50 values were determined with approximately 1 μM for TSA.
Figure 1:In vitro Effects of TSA (0.5, 1, 5, and 10 μM) on the Breast Cancer SK-BR-3 Cells Viability Determined by MTT Assay after 24 and 48 h of Treatment. As shown, from left to the right, the first column of each group belongs to the control group. Other columns belong to TSA treated groups at the concentrations of 0.5, 1, 5, and 10 μM. Values are means of three experiments in triplicate. Asterisks (*) demonstrate significant differences between treated and untreated control groups.
Result of cell apoptosis assay
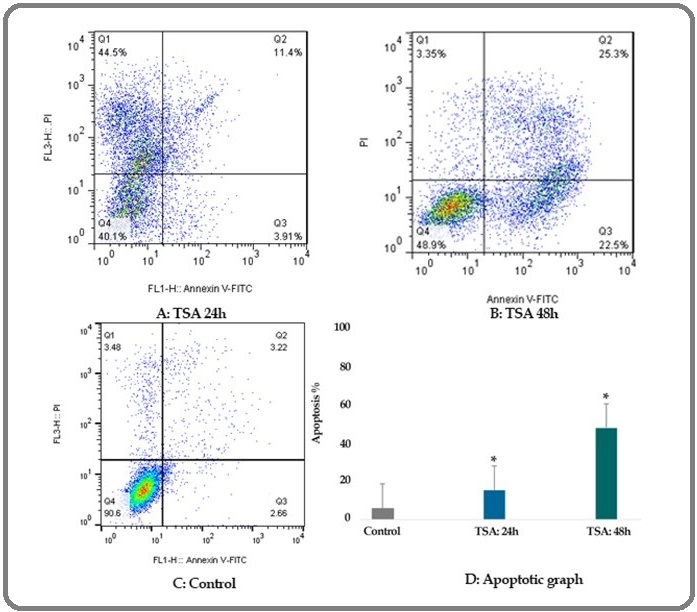
Flow cytometric analysis was done to evaluate the effect of TSA (1 μM) on breast cancer SK-BR-3 cells apoptosis. The percentage of TSA treated and un-treated apoptotic cells was determined by staining with annexin V-FITC and PI after 24 and 48 h of treatment. The apoptosis percentage of the cells was 15.31 and 47.8 (P < 0.001) in different times (24 and 48 h) respectively as shown in Table 2 and Figure 2.
| Cell line | Gene | Drug | Dose (μM) | Duration (h) | Expression | P-value |
| SK-BR-3 | P21 | TSA | 1 | 24 | 1.7 | 0.001 |
| SK-BR-3 | P21 | TSA | 1 | 48 | 2.1 | 0.001 |
| SK-BR-3 | P27 | TSA | 1 | 24 | 1.9 | 0.001 |
| SK-BR-3 | P27 | TSA | 1 | 48 | 2.3 | 0.001 |
| SK-BR-3 | P57 | TSA | 1 | 24 | 2.4 | 0.001 |
| SK-BR-3 | P57 | TSA | 1 | 48 | 2.9 | 0.001 |
| SK-BR-3 | HDAC1 | TSA | 1 | 24 | 0.55 | 0.001 |
| SK-BR-3 | HDAC1 | TSA | 1 | 48 | 0.46 | 0.001 |
| SK-BR-3 | HDAC2 | TSA | 1 | 24 | 0.48 | 0.001 |
| SK-BR-3 | HDAC2 | TSA | 1 | 48 | 0.39 | 0.001 |
| SK-BR-3 | HDAC3 | TSA | 1 | 24 | 0.4 | 0.001 |
| SK-BR-3 | HDAC3 | TSA | 1 | 48 | 0.31 | 0.001 |
Further, maximum apoptosis was seen after 48 h.
Figure 2: The Apoptotic Effect of TSA (1 μM) on Breast Cancer SK-BR-3 Cells Versus Control Groups at Different Periods, 24 and 48h. The SK-BR-3 cells were treated with TSA (1 μM) for 24 and 48h and then the apoptotic effect was assessed by flow cytometric analysis. Results were obtained from three independent experiments and were expressed as mean ± standard error of the mean.
Result of determination of genes expression
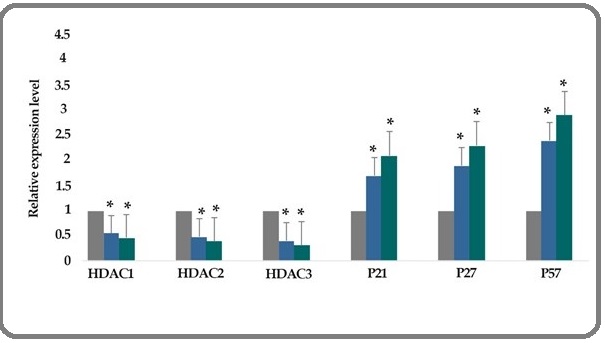
The effect of TSA (1 μM) on the histone deacetylase 1, 2, and 3, p21, p27, and p57 gene expression genes expression was investigated by quantitative real-time RT-PCR analysis. The result indicated that treatment of breast cancer SK-BR-3 cells with TSA (1 μM) for 24 and 48 h downregulated histone deacetylases 1, 2, and 3 and upregulated the p21, p27, and p57 genes significantly, Figure 3.
Figure 3:The Relative Expression Level of Deacetylase 1, 2 and 3, p21, p27, and p57 Gene Expression Genes Expression Treated with TSA (1 μM) Versus Untreated Control Groups at Different Periods, 24 and 48h. The first column of each group belongs to the untreated control group and the others belong to the treated cells with TSA. Asterisks (*) indicate significant differences between the treated and untreated groups.
Discussion
Epigenetic inactivation of TSGs is a hallmark of cancer cells. These epigenetic molecular mechanisms include crosstalk between histone modification, DNA methylation, and other components of chromatin higher-order structure, and lead to the regulation of gene expression. Histone deacetylation plays an important role in the silencing of cancer-related genes, thereby affecting numerous processes, such as the cell cycle checkpoint, signal transduction, apoptosis, angiogenesis, and cell adhesion. Re-expression of TSGs epigenetically inactivated can result in the suppression of cancer cell growth [19]. In the current study, we report that TSA can down-regulate HDAC 1, 2, and 3 and up-regulate p21, p27, and p57 resulting in cell growth inhibition and apoptosis induction in breast cancer SK-BR-3 cell line.
Similarly, it has been reported that HDACs suppresses the growth of breast cancer cell MCF-7, induce apoptosis, and arrested G1 phase of MCF-7 cells by p21 up-regulation and cyclin D1 down-regulation [20]. Several in vitro study have demonstrated that DACIs increased p57Kip2 gene expression in HL-60 (from promyelocytic leukemia); K562 (from an erythroleukemia); HT-29 and CaCO2 (from colon cancers); HeLa (from cervical cancer), Lan-5, SK-N-SH and SK-N-BE (from neuroblastomas); and HEK293 (from the human embryonic kidney) [21]. Meanwhile, other studies have confirmed that the HDACs are the main molecular target for anticancer effect of TSA in breast cancer [22]. In colon cancer, it has been demonstrated that Class I HDACs are involved in repressing p21 and suggest that the growth inhibitory and apoptotic effects induced by HDAC inhibitors are mediated through the inhibition of these enzymes [23]. All reports mentioned above are inconsistent with our findings. Additionally, other researchers have reported various molecular pathways for HDACIs. It has been shown that induction of p21WAF1/Cip1 mediated by silencing of HDAC4 arrests cancer cell growth in human glioblastoma model [24]. In vitro study has been indicated that HDAC4 promotes gastric cancer cell progression mediated through the repression of p21 [25]. In urothelial cancer (HDAC4, HDAC5, and HDAC7) cell lines, treatment with the pan-HDAC inhibitor vorinostat down-regulates the HDAC2 and HDAC8 [26]. Besides, it has been shown that HDACIs can
Down-regulate pro-survival proteins such as Bcl-1 and Bcl-2 which regulate mitochondrial integrity, and up-regulate pro-apoptotic proteins like Bim, Bak, and Bax, which function as sensors of cellular stress and initiate the intrinsic pathway [27]. In summary, HDACIs can play their roles through two molecular mechanisms including intrinsic and extrinsic pathways. In this study, we did not evaluate the intrinsic mechanism. Therefore, this evaluation is recommended.
In conclusion, our findings indicated that the TSA can reactivate the p21, p27, and p57 through down-regulation of deacetylase 1, 2, and 3 gene expression which can open a new window in breast cancer treatment.
Acknowledgments
This article was supported by the adjutancy of research of Jahrom University of Medical Sciences, Iran.
Conflict of interest
The authors report no conflict of interest.
References
- DNA Demethylation and Carcinogenesis Kisseljova N. P., Kisseljov F. L.. Biochemistry (Moscow).2005;70(7). CrossRef
- Histone deacetylases and cancer: causes and therapies Marks Paul A., Rifkind Richard A., Richon Victoria M., Breslow Ronald, Miller Thomas, Kelly William K.. Nature Reviews Cancer.2001;1(3). CrossRef
- HDAC family: What are the cancer relevant targets? Witt Olaf, Deubzer Hedwig E., Milde Till, Oehme Ina. Cancer Letters.2009;277(1). CrossRef
- Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer - overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression Müller Berit Maria, Jana Lisa, Kasajima Atsuko, Lehmann Annika, Prinzler Judith, Budczies Jan, Winzer Klaus-Jürgen, Dietel Manfred, Weichert Wilko, Denkert Carsten. BMC Cancer.2013;13(1). CrossRef
- Class I histone deacetylases 1, 2 and 3 are highly expressed in renal cell cancer Fritzsche FR, Weichert W, Röske A, Gekeler V, Beckers T, Stephan C, Jung K, et a . BMC Cancer.2008;8(1):381.
- Expression of histone deacetylases 1, 2 and 3 in urothelial bladder cancer Poyet Cédric, Jentsch Bastian, Hermanns Thomas, Schweckendiek Daniel, Seifert Hans-Helge, Schmidtpeter Martin, Sulser Tullio, Moch Holger, Wild Peter J, Kristiansen Glen. BMC Clinical Pathology.2014;14(1). CrossRef
- Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy Weichert W, Röske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche F R, Niesporek S, Denkert C, Dietel M, Kristiansen G. British Journal of Cancer.2008;98(3). CrossRef
- Effect of 5-aza-2'-deoxycytidine in comparison to valproic acid and trichostatin a on histone deacetylase 1, dna methyltransferase 1, and cip/kip family (p21, p27, and p57) genes expression, cell growth inhibition, and apoptosis induction in colon cancer sw480 cell line Kavoosi Fraidoon, Sanaei Masumeh. Advanced Biomedical Research.2019;8(1). CrossRef
- Histone Deacetylase Inhibitors in Clinical Studies as Templates for New Anticancer Agents Mottamal Madhusoodanan, Zheng Shilong, Huang Tien, Wang Guangdi. Molecules.2015;20(3). CrossRef
- Trichostatin A causes p53 to switch oxidative-damaged colorectal cancer cells from cell cycle arrest into apoptosis Habold C., Poehlmann A., Bajbouj K., Hartig R., Korkmaz K.S., Roessner A., Schneider-Stock R.. Journal of Cellular and Molecular Medicine.2008;12(2). CrossRef
- Effect of valproic acid in comparison with vorinostat on cell growth inhibition and apoptosis induction in the human colon cancer SW48 cells in vitro. Exp Oncol 2018 Sanaei M, Kavoosi F, Mansoori O. .
- Effect of curcumin and trichostatin a on the expression of DNA methyltransfrase 1 in hepatocellular carcinoma cell line hepa 1-6 Kavoosi F. Iranian Journal of Pediatric Hematology and Oncology.2018;8(4):193-201.
- Comparative analysis of the effects of valproic acid and tamoxifen on proliferation, and apoptosis of human hepatocellular carcinoma WCH 17 celllin Kavoosi F, Sanaei M. Iranian Journal of Pediatric Hematology and Oncology.2018;8(1):12-20.
- Effects of 5-aza-2ˈ-deoxycytidine and Valproic Acid on Epigenetic-modifying DNMT1 Gene Expression, Apoptosis Induction and Cell Viability in Hepatocellular Carcinoma WCH-17 cell line Sanaei M, Kavoosi F. Iranian Journal of Pediatric Hematology & Oncology 2019..
- Effect of Genistein in Comparison with Trichostatin A on Reactivation of DNMTs Genes in Hepatocellular Carcinoma Sanaei Masumeh, Kavoosi Fraidoon, Roustazadeh Abazar, Golestan Fatemeh. Journal of Clinical and Translational Hepatology.2018;6(2). CrossRef
- Effect of 5-aza-2'-deoxycytidine in comparison to valproic acid and trichostatin a on histone deacetylase 1, dna methyltransferase 1, and cip/kip family (p21, p27, and p57) genes expression, cell growth inhibition, and apoptosis induction in colon cancer sw480 cell line Kavoosi Fraidoon, Sanaei Masumeh. Advanced Biomedical Research.2019;8(1). CrossRef
- Expression profile of histone deacetylases 1, 2 and 3 in ovarian cancer tissues Jin Ke Long, Pak Jhang Ho, Park Jeong-Yeol, Choi Won Ho, Lee Joo-Yong, Kim Jong-Hyeok, Nam Joo-Hyun. Journal of Gynecologic Oncology.2008;19(3). CrossRef
- MicroRNA-222 regulates MMP-13 via targeting HDAC-4 during osteoarthritis pathogenesis Song Jinsoo, Jin Eun-Heui, Kim Dongkyun, Kim Keun Young, Chun Churl-Hong, Jin Eun-Jung. BBA Clinical.2015;3. CrossRef
- Epigenomics and epigenetic therapy of cancer Brown Robert, Strathdee Gordon. Trends in Molecular Medicine.2002;8(4). CrossRef
- The role and possible molecular mechanism of valproic acid in the growth of MCF-7 breast cancer cells Ma Xiao-jie, Wang Yun-shan, Gu Wei-ping, Zhao Xia. Croatian Medical Journal.2017;58(5). CrossRef
- Histone deacetylase inhibitors upregulate p57Kip2 level by enhancing its expression through Sp1 transcription factor Cucciolla V., Borriello A., Criscuolo M., Sinisi A. A., Bencivenga D., Tramontano A., Scudieri A. C., Oliva A., Zappia V., Ragione F. D.. Carcinogenesis.2007;29(3). CrossRef
- Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo Vigushin DM, Ali S, Pace PE, Mirsaidi N, Ito K, Adcock I, et al . Clin Cancer Res.2001;7(4):971-976.
- Histone Deacetylase 3 (HDAC3) and Other Class I HDACs Regulate Colon Cell Maturation and p21 Expression and Are Deregulated in Human Colon Cancer Wilson Andrew J., Byun Do-Sun, Popova Natalia, Murray Lucas B., L'Italien Kaitlin, Sowa Yoshihiro, Arango Diego, Velcich Anna, Augenlicht Leonard H., Mariadason John M.. Journal of Biological Chemistry.2006;281(19). CrossRef
- HDAC4 represses p21WAF1/Cip1 expression in human cancer cells through a Sp1-dependent, p53-independent mechanism Mottet D, Pirotte S, Lamour V, Hagedorn M, Javerzat S, Bikfalvi A, Bellahcène A, Verdin E, Castronovo V. Oncogene.2008;28(2). CrossRef
- Histone Deacetylase HDAC4 Promotes Gastric Cancer SGC-7901 Cells Progression via p21 Repression Kang Zhen-Hua, Wang Chun-Yan, Zhang Wen-Liang, Zhang Jian-Tao, Yuan Chun-Hua, Zhao Ping-Wei, Lin Yu-Yang, Hong Sen, Li Chen-Yao, Wang Lei. PLoS ONE.2014;9(6). CrossRef
- Changes in histone deacetylase (HDAC) expression patterns and activity of HDAC inhibitors in urothelial cancers Niegisch Günter, Knievel Judith, Koch Annemarie, Hader Christiane, Fischer Ute, Albers Peter, Schulz Wolfgang A.. Urologic Oncology: Seminars and Original Investigations.2013;31(8). CrossRef
- Vorinostat-An overview Bubna AdityaKumar. Indian Journal of Dermatology.2015;60(4). CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2020
Author Details