PIK3CA, KI67, Estrogen (ER) and Progesterone Receptors (PR) Expression Pattern of in HER2 Positive Breast Cancers
Download
Abstract
Background: PIK3CA mutations have been reported to be associated with resistance to therapy in HER2+ breast cancers. This study, therefore, became imperative to determine the expression pattern of this mutant protein together with ER, PR and KI67 in order to serve as a useful predictive tool in the management of HER2 breast cancers.
Methods: A total of 53 archived formalin-fixed, paraffin-embedded HER2+ breast cancer tissue blocks from 2015 to 2019 were used for the study in NAUTH Nnewi. The selected blocks were sectioned and stained with haematoxylin and eosin staining techniques. HER2, ER and PR status confirmation as well as PIK3CA and KI67 protein expressions were evaluated using immunohistochemistry (Avidin-biotin complex method). PIK3CA and KI67 expressions in the tissue were scored based on proportion and intensity of immune-labelling using the semi-quantitative method.
Result: The mean age of subjects was 47 years and the breast cancers were all invasive ductal carcinoma. Twenty-nine (54.7%) were ER+ while 24 (45.3%) were ER-. Twenty-one (39.6%) were PR+ while 32 (60.4%) were PR-. Twenty-one (39.6%) were PIK3CA negative, 9(35.8%) showed low PIK3CA, while 13(24.5%) showed high PIK3CA. Thirty-four (64.2%) were negative for KI67, 11(20.8%) showed low KI67, while 8(15.5%) showed high KI67. There was weak and moderate positive relationship between ER/PR status and PIK3CA (r=-0.032; p=0.822) and KI67 (r=0.050; p=0.721) respectively. A weak negative correlation between KI67 and PIK3CA (r=-0.118; p=0.401) were observed with 12 (22.4%) of the 13 highly positive PIK3CA cases showing either negative or low for KI67 immunoreactivity while 7(13.2%) of the 8 highly positive KI67 cases showed either negative or low PIK3CA immunoreactivity.
Conclusion: This study established a moderate expression of PIK3CA mutant protein. It also pointed out an existing interesting relationship between PIK3CA and KI67, which can be further revealed in future studies.
Introduction
Female breast cancer is currently the leading cause of global cancer incidence, with an estimated 2.3 million new cases, representing 11.7% of all cancer cases [1]. It is the fifth leading cause of cancer mortality worldwide, with 685,000 deaths [1]. Among women, breast cancer accounts for 1 in 4 cancer cases and for 1 in 6 cancer deaths, ranking first for incidence in the vast majority of countries and for mortality in 110 countries [1]. Nigeria is the most populous country in Africa, with approximately 20% of the continent’s population and slightly more than half that of West Africa [2, 3]. According to Global Cancer Statistics 2020, the incidence of breast cancer in Nigeria is 22.7 per 100,000 women and its age standardized mortality ratio is 18.1 per 100,000 women and the 5year prevalence of breast cancer in Nigeria is 59.31 [1]. Among other factors, the growing cases of breast cancer in Nigeria seemed to have been worsened by the steeply increasing size of the population at risk [4]. Clinically, breast cancers are categorized into subtypes, to facilitate targeted therapy, using the three standard immunohistochemical markers, viz; ‘luminal A’ (ER+ and/or PR+ & HER2-), ‘luminal B’ (ER+and/or PR+, HER2+), ‘HER2 overexpressing’ (ER-, PR-, HER2-) [5]. HER2 overexpressing breast cancer is an aggressive form of the disease and about one in five women diagnosed with breast cancer worldwide will have HER2-positive breast cancer [6]. The natural history and prognosis of breast cancer cells expressing high levels of HER2 is associated with more aggressive tumors and poor sensitivity to standard chemotherapeutic agents [7].
Although Trastuzumab, a humanized monoclonal antibody to the HER2 protein, has shown efficacy in patients whose tumors are HER2+[8], however, the overall response rate to trastuzumab is low, and almost half of patients with HER2-positive breast cancer exhibit a primary resistance to trastuzumab-based therapy [9, 10, 11]. Resistance to trastuzumab, is a major clinical concern facing breast oncologists today. This prompted research into the mechanisms of Trastuzumab resistance, which are thought to underlie failure of therapy of which PIK3CA mutation, reported to be present in over one-third of breast cancer cases [12], has been highly implicated [13, 14, 15, 16]. PIK3CA mutation frequency varies among breast cancer subtypes and among different geographical locations and has recently been found to be increasing. While several testing methods and chemotherapeutic agents have been developed to identify and target the phosphoinositide 3-kinase (PI3K) pathway, the therascreen PIK3CA mutation assay and the alpha-specific PI3K inhibitor alpelisib have been approved by Food and Drug Administration (FDA) for identifying and treating patients with advanced PIK3CA-mutated breast cancer. In Nigeria so far, HER2 positive breast cancer patients are placed on trastuzumab therapy without testing their PIK3CA mutation status despite the fact that the PIK3CA mutation assay and the alpha-specific PI3K inhibitor alpelisib have been approved by Food and Drug Administration (FDA) for identifying and treating patients with advanced PIK3CA-mutated breast cancer [17]. This study was therefore, aimed at evaluating PIK3CA and KI67 immunoreactivity pattern as predictive and therapeutic biomarkers in HER2 positive breast cancers. While several studies have been previously conducted to determine expression patterns of PIK3CA and KI67 in breast cancers, in Nigeria so far, there is no data available about PIK3CA mutations frequency in HER-2 positive breast cancer patients, neither is there any research that considered relating PIK3CA with the proliferative marker KI67; these gap in knowledge, when bridged could be of help in future research and in clinical application of PIK3CA as a predictive marker for anti-HER2 therapy. Determining the expression pattern of PIK3CA in HER2 positive breast cancer patients before treatment is commenced will help to identify those who will likely have primary resistance to trastuzumab-based therapy and are most likely to benefit from treatment using a PI3K inhibitor; and relating the PIK3CA expression pattern with the proliferative marker KI67 will help identify the proliferative pattern of mutant HER2 positive breast cancers. This study therefore becomes imperative to evaluate PIK3CA and KI67 immunoreactivity in HER2 positive breast cancers. Since it has been established in the literature from previous studies that almost half of patients with HER2-positive breast cancer exhibit a primary resistance to trastuzumab-based therapy, rather than subjecting these patients to the various side effects of trastuzumab-based therapy which they may not benefit from, assessment of PIK3CA and KI67 immuno- reactivities could be a promising good combination for prediction of treatment response in HER2 positive breast cancer patients and can help guide to a more timely and adequate treatment plan for these group of patients. Due to the present challenge of resistance to therapy among HER2 positive breast cancer patients, which has been linked to PIK3CA mutations, the mutation which when present in tumors has been stated to result to increase in proliferation of the tumor cells (increased ki67 index), this study therefore, evaluated the immunoraectivity pattern of PIK3CA and KI67 in HER2 positive breast cancers.
Materials and Methods
Study design
This was a retrospective cross sectional study carried out at Nnamdi Azikiwe University Teaching Hospital (NAUTH) Nnewi, to determine the association of PIK3CA, KI67, ER and PR in HER2 positive breast cancers. The research proposal was submitted to Ethics Research Committee of the hospital for consideration and an approval was obtained with reference number (NAUTH/ CS/66/VOL.12/224/2019/087), before commencement of the study.
Tissue samples
A total of 273 breast cancer cases comprising of 65 HER2 positive and 208 HER 2 negative cases were recorded from 2015-2019. Of the 65 cases of HER2 positive breast cancer cases, 10 cases were excluded either because of unavailable clinical data or because of insufficient tissues in the paraffin blocks. Therefore, 53 formalin-fixed, paraffin wax-embedded archived tissue blocks of HER2 positive breast cancers were selected for the study while 4 noncancerous breast tissue specimens were used as control.
Tissue blocks and the demographic data of the subjects were retrieved from the archive of Histopathology department of the hospital. Selected tissue blocks were trimmed and sectioned at 3-micron (3µ) thick sections on a rotary microtome (HM340E Thermo Scientific. Massachusetts, United States of America). The sections were divided into six (6) groups: first group was stained using Haematoxylin and Eosin staining technique for confirmation of diagnosis, second group was stained for HER2,using HER2 monoclonal antibody kit for confirmation of HER2 positivity while third, fourth, fifth and sixth groups were stained for ER, PR, PIK3CA and KI67 respectively, using Avidin-biotin immunohistochemistry (IHC) method. Archived formalin fixed paraffin embedded normal breast tissues were used for study control, while the manufacturer in the purchased respective antibody kits included IHC control.
Haematoxylin and Eosin Staining Technique [18]
Sections were stained by haematoxylin and eosin staining technique and micrographs were taken using Olympus microscope (BHTU. New York Microscope Company). The stained slides first underwent independent blind reviews to reconfirm the diagnosis of breast cancer. There were diagnostic agreement between the two reviewers, and so consensus diagnosis was achieved and no additional reviews were conducted.
Immunohistochemistry (IHC) Staining Technique [19]
The slides were stained using the Avidin-Biotin complex immunohistochemistry method. Monoclonal antibodies of HER2, ER, PR, PIK3CA and KI67 were employed. Exposed Mouse and Rabbit Specific HRP/DAB detection IHC kit were employed for immunostaining while detection of immunoreactivity was performed according to manufacturer’s instruction. Both antibodies and detection kits were procured from Abcam Plc Cambridge UK. Stained sections were reviewed for nuclear immunoreactivity.
Negative expression was recorded when none of the tumor nuclei stained for either HER2, PIK3CA or KI67 showed nuclear staining and protein expression was recorded as positive if both the tumor and internal control showed nuclear staining. Those tumors showing negative of expression for HER2, ER, PR, PIK3CA or KI67 in all tumor nuclei were regarded as negative, and those tumors with positive expression for ER, PR, PIK3CA and KI67 were scored and described as either high or low. Micrographs were taken using Olympus microscope.
Immunohistochemistry Scoring [20]
The immunohistochemical scoring analysis was performed using semiquantitative scoring method. At least 5 high power fields were evaluated for each tumour and the staining rate of the tumour cells were calculated. Amean percentage of stained tumour cells was determined and graded as follows; negative indicated no stained tumour cell, immunopositivity: weak for 1-10% positive tumour cells, moderate for10-50% positive tumour cells, strong for 51-100% positive tumour cells.
Data Analysis
Data analysis were carried out with the aid of SPSS version 22. Associations between variables were tested using Spearman’s correlation analysis. P<0.05 was considered significant.
Results
Immunoreactivity Pattern of PIK3CA, KI67, ER and PR
The immunoreactivity patterns of estrogen (ER) and progesterone receptors (PR) in HER2 positive breast cancers showed 29 (54.7%) and 21 (39.6%) immunopositivity, and 24 (45.3%) and 32 (60.4%) negative staining for ER and PR respectively. Similarly, PIK3CA protein revealed 21 (39.6%) negative staining, 19 (35.8%) low positive staining and 13 (24.5%) high positive staining while KI67 revealed 34 (64.2%) negative staining, 11 (20.8%) low immunostaining and 8 (15.5%) high immunostaining (Table 1).
| Immunoreactivity Pattern | Estrogen Receptor (ER) | Progesterone Receptor (PR) | PIK3CA | KI67 |
| Negative | 29 (54.7%) | 32 (60.4%) | 21 (39.6%) | 34 (64.2%) |
| Low Positive | 24 (45.3%) | 21 (39.6%) | 19 (35.8%) | 11 (20.8%) |
| High Positive | - | - | 13 (24.5%) | 8 (15.5%) |
| Total | 53 (100%) | 53 (100%) | 53 (100%) | 53 (100%) |
Correlations of ER/PR Status with PIK3CA and KI67 Proteins expression
The correlation of ER/PR status with PIK3CA protein expression and KI67 protein expression in the HER2 positive breast cancers are as follows. There was no significant relationship between ER/PR status and PIK3CA protein expression (r=-0.032; p=0.822). Of the 13 cases that showed high PIK3CA protein expression, 7 were hormone receptor positive while 6 were hormone receptor negative. There was a non significant weak positive correlation (r=0.050; p=0.721) between ER/ PR status and KI67 expression. Out of the 8 cases that had high KI67 indices, 5 (9.4%) were hormone receptor positive while 3 (5.6%) were hormone receptor negative. There was a weak negative correlation (r=-0.118) between ki67protein expression and Pik3CA protein expression, though the data are not significant (p=0.401). Out of the 13 high cases of PIK3CA protein expression, 12 (22.4%) corresponded with negative/low KI67 protein expression, and out of the 8 high cases of KI67 protein expression, 7 (13.2%) corresponded with negative/low PIK3CA protein expression (Table 2).
| ER/PR | KI67 | PIK3CA | ||
| Correlation coefficient | 1.000 | 0.05 | -0.032 | |
| ER/PR | Sig. (2-tailed) | 0.721 | 0.822 | |
| N | 53 | 53 | 53 | |
| Correlation coefficient | 0.05 | 1.000 | -0.118 | |
| KI67 | Sig. (2-tailed) | 0.721 | 0.401 | |
| N | 53 | 53 | 53 | |
| Correlation coefficient | -0.03 | -0.118 | 1.000 | |
| PIK3CA | Sig. (2-tailed) | 0.822 | 0.401 | |
| N | 53 | 53 | 53 |
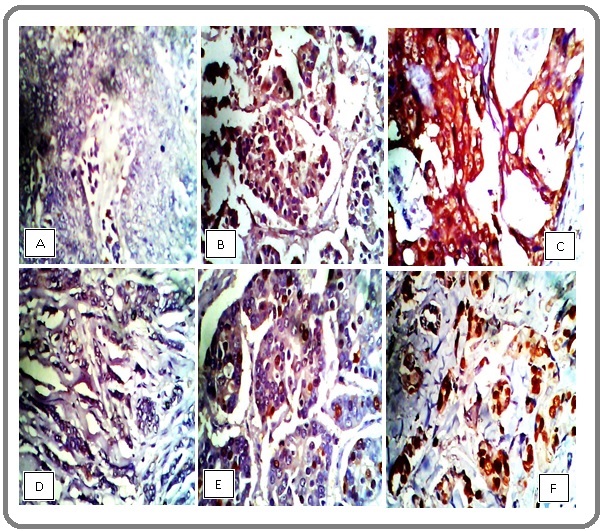
Immunohistochemical staining micrographs of breast tissues
PlateI is representative photomicrographs of the stained slides from the current study. Showing from left to right; (A) negative, (B) low and (C) high expression of PIK3CA protein and KI67 (D) negative, (E) low and (F) high expressions in HER2 positive cancers. The staining patterns were identified by the negative, mild and intense brown colorations of the nuclei of the cancer cells (Figure 1).
Figure 1. Plate I, Immunohistochemical Staining of Breast Tissue, Showing from Left to Right; (A) Negative, (B) Low and (C) High Expression of PIK3CA Protein and KI67 (D) Negative, (E) Low and (F) High Expressions in a HER2 Positive Cancers. X400 Magnifications.
Discussion
The findings of the study showedHER2 positive cancers with higher percentage of ER positive tumors than were PR positive tumors. This corroborates the findings of Elwy et al [21] who also had more ER positive cases compared to PR positive cases. This is quite understandable because ER positive tumors are the most prevalent of all the molecular subtypes of breast cancer. Contrarily, PIK3CA immunoreactive cancers had more positive PR staining than ER. This however, did not agree with the report of Allison [22], who observed and reported that ER positive tumors tend to harbor more severe genetic aberrations when compared to PR positive tumors. Olivotto et al [23], in a related study reported that PR positive tumors are hardly ER negative, which agrees with the finding of this study, as only on one instance did we have a PR positive tumor that was ER negative. The low percentage of ER positive tumors with PR negative disagrees partly with the observation Rakha et al [24], who found approximately 40% ER positive tumors to be PR negative. Similarly, the percentages of bi-positive (35.8%) (ER+PR+) and bi-negative cancers in the present study did not agree with that earlier reports of Rakhaet al [24] and Dunnwald et al [25] but partly corroborated the reports of Elledge et al [26] and Bardou et al [27]. The current report could probably, be due to the fact that the current study was limited to only HER2 positive breast cancers while Rakha et al [24] in their study included all breast cancer types. The disagreement could attributable to variations in age, geographical location and the scope of the subjects involved.
The expression pattern of PIK3CA protein as reported in the present study (24.5%) disagrees with a related study by Elwy et al [21], Lian et al [28], Aziz et al [29] and Shimoi et al [31]. Elwy et al. (2017) [22] and Lian et al. (2020) [28], reported low frequency of 3.7% and 15.7% respectively while Aziz et al [29] and Shimoi et al [30] reported higher expression frequencies of 45% and 33% respectively. The variations in expression frequency could be explained by the different geographical locations and the breast cancer subtypes in the various studies involved. The importance of the finding lies in the fact that PIK3CA protein was over-expressed in certain kinds of breast cancers and this should for an insight when planning treatment regimen for individual patients. This also, suggests that PIK3CA mutations (expression) in breast cancer vary among different breast cancer subtypes and different geographical locations. A lesser percentage 8/53 (15.0%) of the tumors in the current study expressing Ki67 protein disagrees with the finding of Yerushalmi et al [31] who reported that HER2 positive tumors are usually associated with high KI67 expression. The high frequent negative and low KI67 expression pattern may suggest less aggressive tumors and better prognosis, but could also be a function of geographical location and cancer sub-type. A non-significant moderate positive relationship exist between ER/PR status and PIK3CA protein expression (r=-0.032; p=0.822). This corroborates previous studies by Lian et al [28] and Elwy et al [21] who found no significant correlation between PIK3CA and ER/PR expression. The present study reported that13% ER/PR positive and 11.3% ER/PR negative cancers were highly positive for PIK3CA respectively. This is in line with the findings of Ahmad et al [32]; Hu et al [33] and Wu et al [34], who found that the PIK3CA somatic mutations in hormonal positive/HER2 positive breast cancers are more common, though the data were not significant. This however, partly disagrees with the earlier report by Arsenic et al [35], where the PIK3CA mutations rate was higher in ER+PR+/ HER2 (-) cancers compared to ER+PR+/HER2 (+) cases and those of Elwy et al [21] who observed that ER-/PR- breast cancer pattern had more PIK3CA mutation when compared with ER+/PR+ pattern. There seems to be an ill-defined relationship between hormone secreting breast cancers and non-hormone secreting ones, which cannot be fully elucidated in the current study. The study observed that hormone secreting breast cancers slightly express more PIK3CA than non hormone secreting breast cancers, which, however, calls for further studies.
There was a moderate positive correlation (r=0.050) between ER/PR status and KI67 expression, but the data are not significant (p=0.721). A greater percentage 5/8 (9.4%) of the tumors with high KI67 immunoreactivity, were hormone receptor positive while a lesser percentage 3/8 (5.6%) were hormone receptor negative. This is contrary to the observation made by Yerushalmi et al [31] who reported high KI67 index is associated with hormone receptor negative status, but in line with previous studies by Parise and Caggiano [36] and Howlader et al [37] who reported that luminal B breast cancers (HR+/HER2+) are defined by being highly positive for the protein ki67. Whereas the present study observed greater number of the high KI67 index cases were hormone receptor positive, the percentage may not be enough to conclude that hormone-secreting breast cancers were defined by being highly positive for the protein KI67, however, the relationship should be elucidated further.
The study further observed a weak negative correlation (r=-0.118) between KI67protein expression and PIK3CA protein expression, though the data were not statistically significant (p=0.401). This suggests that high PIK3CA protein expression suppresses cellular proliferation.
This seemingly interesting finding looks rather like a contradiction of an established fact that links PIK3CA protein activation with cellular proliferation in tumors. This observation however, may not be unconnected to a unique character of mutant HER2 positive breast cancers. The PIK3CA expression therefore, rather conferring disadvantage to breast cancer patients expressing them(because of the resistance to chemotherapy which it is known to cause), potentially promises to be an advantage to HER2 positive breast cancers by decreasing the proliferation rate of the cancer cells. Recall that, according to Denkert et al [38], low proliferating tumors, even when not responding to chemotherapy have good prognosis (low Ki67 linked to good outcome) whereas, high proliferating tumors that are chemotherapy or hormone therapy resistant have poor prognosis (high Ki67 linked to poor outcome). It is noteworthy, however, that although, this finding looks interesting and promising, the relationship is weak and therefore, requires further studies to provide more insight. Summarily, to the best of our knowledge, the present study is the first one which investigated PIK3CA mutations in HER2-positive breast cancers in Nigeria and the first to investigate the relationship between PIK3CA and KI67 in these subjects. The PIK3CA expression frequency obtained in the present study (24.5%) is moderate compared to the frequencies of previous studies. A greater percentage of the tumors (85%) were either negative or low for the proliferative marker KI67expression. There was no significant relationship between ER/PR status and PIK3CA or between ER/PR status and KI67. The current study revealed a non-significant inverse interesting relationship between PIK3CA and KI67. In conclusion, the knowledge of pretreatment PIK3CA mutation status and KI67 index of HER2 positive breast cancers potentially favors HER2 positive breast cancers as evident in the low or negative KI67 immunoreactivity in high PIK3CA immunoreactive breast cancers. This indicated a low tumor proliferation rate in PIK3CA mutant HER2 positive breast cancers. Further studies, with larger sample sizes to quantitative and unravel the mutation sequence will be undertaken to reveal the actual relationship between PIK3CA and KI67 proteins expression.
Acknowledgements
The authors wish to express their immense gratitude the staff and management of the department of Histopathology, NnamdiAzikiwe University Teaching Hospital Nnewi for granting us the approval to carry out the study and to Professor C. O. Akosile for proof reading the manuscript.
Funding statement
This research received no specific grant from any funding agency in the public, commercial, or not-for- profit sectors.
References
- Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries Sung Hyuna, Ferlay Jacques, Siegel Rebecca L., Laversanne Mathieu, Soerjomataram Isabelle, Jemal Ahmedin, Bray Freddie. CA: a cancer journal for clinicians.2021;71(3). CrossRef
- Cancer incidence in Nigeria: a report from population-based cancer registries Jedy-Agba Elima, Curado Maria Paula, Ogunbiyi Olufemi, Oga Emmanuel, Fabowale Toyin, Igbinoba Festus, Osubor Gloria, Otu Theresa, Kumai Henry, Koechlin Alice, Osinubi Patience, Dakum Patrick, Blattner William, Adebamowo Clement A.. Cancer Epidemiology.2012;36(5). CrossRef
- A million africans a year dying from cancer by 2030: what can cancer research and control offer to the continent? Sylla Bakary S., Wild Christopher P.. International Journal of Cancer.2012;130(2). CrossRef
- Emerging breast cancer epidemic: evidence from Africa Akarolo-Anthony Sally N., Ogundiran Temidayo O., Adebamowo Clement A.. Breast cancer research: BCR.2010;12 Suppl 4. CrossRef
- Ultrasonographic features of triple-negative breast cancer: a comparison with other breast cancer subtypes Yang Qi, Liu Hong-Yan, Liu Dan, Song Yan-Qiu. Asian Pacific journal of cancer prevention: APJCP.2015;16(8). CrossRef
- Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update Wolff Antonio C., Hammond M. Elizabeth H., Hicks David G., Dowsett Mitch, McShane Lisa M., Allison Kimberly H., Allred Donald C., Bartlett John M. S., Bilous Michael, Fitzgibbons Patrick, Hanna Wedad, Jenkins Robert B., Mangu Pamela B., Paik Soonmyung, Perez Edith A., Press Michael F., Spears Patricia A., Vance Gail H., Viale Giuseppe, Hayes Daniel F.. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology.2013;31(31). CrossRef
- Emerging Therapeutic Options for HER2-Positive Breast Cancer Martin Miguel, López-Tarruella Sara. American Society of Clinical Oncology Educational Book. American Society of Clinical Oncology. Annual Meeting.2016;35. CrossRef
- Effect of hormone replacement therapy on breast cancer risk: estrogen versus estrogen plus progestin Ross R. K., Paganini-Hill A., Wan P. C., Pike M. C.. Journal of the National Cancer Institute.2000;92(4). CrossRef
- PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients Nagata Yoichi, Lan Keng-Hsueh, Zhou Xiaoyan, Tan Ming, Esteva Francisco J., Sahin Aysegul A., Klos Kristine S., Li Ping, Monia Brett P., Nguyen Nina T., Hortobagyi Gabriel N., Hung Mien-Chie, Yu Dihua. Cancer Cell.2004;6(2). CrossRef
- HER-2-positive metastatic breast cancer: trastuzumab and beyond Metro Giulio, Mottolese Marcella, Fabi Alessandra. Expert Opinion on Pharmacotherapy.2008;9(15). CrossRef
- HER2 genomic amplification in circulating tumor DNA and estrogen receptor positivity predict primary resistance to trastuzumab emtansine (T-DM1) in patients with HER2-positive metastatic breast cancer Sakai Hitomi, Tsurutani Junji, Iwasa Tsutomu, Komoike Yoshifumi, Sakai Kazuko, Nishio Kazuto, Nakagawa Kazuhiko. Breast Cancer (Tokyo, Japan).2018;25(5). CrossRef
- PIK3CA mutations in breast cancer: reconciling findings from preclinical and clinical data Zardavas Dimitrios, Phillips Wayne A., Loi Sherene. Breast cancer research: BCR.2014;16(1). CrossRef
- A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer Berns Katrien, Horlings Hugo M., Hennessy Bryan T., Madiredjo Mandy, Hijmans E. Marielle, Beelen Karin, Linn Sabine C., Gonzalez-Angulo Ana Maria, Stemke-Hale Katherine, Hauptmann Michael, Beijersbergen Roderick L., Mills Gordon B., Vijver Marc J., Bernards René. Cancer Cell.2007;12(4). CrossRef
- Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines Kataoka Y., Mukohara T., Shimada H., Saijo N., Hirai M., Minami H.. Annals of Oncology: Official Journal of the European Society for Medical Oncology.2010;21(2). CrossRef
- Low PTEN levels and PIK3CA mutations predict resistance to neoadjuvant lapatinib and trastuzumab without chemotherapy in patients with HER2 over-expressing breast cancer Rimawi Mothaffar F., De Angelis Carmine, Contreras Alejandro, Pareja Fresia, Geyer Felipe C., Burke Kathleen A., Herrera Sabrina, Wang Tao, Mayer Ingrid A., Forero Andres, Nanda Rita, Goetz Matthew P., Chang Jenny C., Krop Ian E., Wolff Antonio C., Pavlick Anne C., Fuqua Suzanne A. W., Gutierrez Carolina, Hilsenbeck Susan G., Li Marilyn M., Weigelt Britta, Reis-Filho Jorge S., Kent Osborne C., Schiff Rachel. Breast Cancer Research and Treatment.2018;167(3). CrossRef
- Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer Vogel Charles L., Cobleigh Melody A., Tripathy Debu, Gutheil John C., Harris Lyndsay N., Fehrenbacher Louis, Slamon Dennis J., Murphy Maureen, Novotny William F., Burchmore Michael, Shak Steven, Stewart Stanford J., Press Michael. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology.2002;20(3). CrossRef
- Frequency and spectrum of PIK3CA somatic mutations in breast cancer Martínez-Sáez Olga, Chic Nuria, Pascual Tomás, Adamo Barbara, Vidal Maria, González-Farré Blanca, Sanfeliu Esther, Schettini Francesco, Conte Benedetta, Brasó-Maristany Fara, Rodríguez Adela, Martínez Débora, Galván Patricia, Rodríguez Ana Belén, Martinez Antonio, Muñoz Montserrat, Prat Aleix. Breast cancer research: BCR.2020;22(1). CrossRef
- Medical Laboratory Science: Theory and Practice. Tata McGraw-Hill Publishing Company Limited, New Delhi, pp480 Ochei J, Kolhatkar A. 2007.
- Diagnostic immunohistochemistry, theranostic and genomic application, 4th edition, Saunders, Philadelphia, pp 520-35 Dabbs DJ, Thompson LRD. 2013.
- Selecting immunohistochemical cut-off scores for novel biomarkers of progression and survival in colorectal cancer Zlobec Inti, Steele Russell, Terracciano Luigi, Jass Jeremy R., Lugli Alessandro. Journal of Clinical Pathology.2007;60(10). CrossRef
- PIK3CA mutations in HER2-positive Breast Cancer Patients; Frequency and Clinicopathological Perspective in Egyptian Patients Elwy Fatma, Helwa Reham, El Leithy Asmaa A., Shehab El din Zeinab, Assem Magda M., Hassan Nagwa H. A.. Asian Pacific journal of cancer prevention: APJCP.2017;18(1). CrossRef
- Molecular pathology of breast cancer: what a pathologist needs to know Allison Kimberly H.. American Journal of Clinical Pathology.2012;138(6). CrossRef
- Time to stop progesterone receptor testing in breast cancer management Olivotto Ivo A., Truong Pauline T., Speers Caroline H., Bernstein Vanessa, Allan Sharon J., Kelly Seamus J., Lesperance Mary L.. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology.2004;22(9). CrossRef
- Biologic and clinical characteristics of breast cancer with single hormone receptor positive phenotype Rakha Emad A., El-Sayed Maysa E., Green Andrew R., Paish E. Claire, Powe Desmond G., Gee Julia, Nicholson Robert I., Lee Andrew H. S., Robertson John F. R., Ellis Ian O.. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology.2007;25(30). CrossRef
- Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients Dunnwald Lisa K., Rossing Mary Anne, Li Christopher I.. Breast cancer research: BCR.2007;9(1). CrossRef
- Estrogen receptor (ER) and progesterone receptor (PgR), by ligand-binding assay compared with ER, PgR and pS2, by immuno-histochemistry in predicting response to tamoxifen in metastatic breast cancer: a Southwest Oncology Group Study Elledge RM, Green S, Pugh R, et al . Int J Cancer.2000;89(2):111-117.
- Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases Bardou Valerie-Jeanne, Arpino Grazia, Elledge Richard M., Osborne C. Kent, Clark Gary M.. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology.2003;21(10). CrossRef
- Clinical-Pathologic Analysis of Breast Cancer With PIK3CA Mutations in Chinese Women Lian Jing, Xu En-Wei, Xi Yan-Feng, Wang Hui-Wen, Bu Peng, Wang Jin-Fen, Wang Li-Xia. Technology in Cancer Research & Treatment.2020;19. CrossRef
- The High Frequency of PIK3CA Mutations in Iranian Breast Cancer Patients Azizi Tabesh Ghasem, Izadi Pantea, Fereidooni Forouzandeh, Emami Razavi Amir Nader, Tavakkoly Bazzaz Javad. Cancer Investigation.2017;35(1). CrossRef
- PIK3CA mutation profiling in patients with breast cancer, using a highly sensitive detection system Shimoi Tatsunori, Hamada Akinobu, Yamagishi Marifu, Hirai Mitsuharu, Yoshida Masayuki, Nishikawa Tadaaki, Sudo Kazuki, Shimomura Akihiko, Noguchi Emi, Yunokawa Mayu, Yonemori Kan, Shimizu Chikako, Kinoshita Takayuki, Fukuda Takahiro, Fujiwara Yasuhiro, Tamura Kenji. Cancer Science.2018;109(8). CrossRef
- Ki67 in breast cancer: prognostic and predictive potential Yerushalmi Rinat, Woods Ryan, Ravdin Peter M., Hayes Malcolm M., Gelmon Karen A.. The Lancet. Oncology.2010;11(2). CrossRef
- Molecular evaluation of PIK3CA gene mutation in breast cancer: determination of frequency, distribution pattern and its association with clinicopathological findings in Indian patients Ahmad Firoz, Badwe Anuya, Verma Geeta, Bhatia Simi, Das Bibhu Ranjan. Medical Oncology (Northwood, London, England).2016;33(7). CrossRef
- Identifying Circulating Tumor DNA Mutation Profiles in Metastatic Breast Cancer Patients with Multiline Resistance Hu Zhe-Yu, Xie Ning, Tian Can, Yang Xiaohong, Liu Liping, Li Jing, Xiao Huawu, Wu Hui, Lu Jun, Gao Jianxiang, Hu Xuming, Cao Min, Shui Zhengrong, Xiao Mengjia, Tang Yu, He Qiongzhi, Chang Lianpeng, Xia Xuefeng, Yi Xin, Liao Qianjin, Ouyang Quchang. EBioMedicine.2018;32. CrossRef
- The distinct clinicopathological and prognostic implications of PIK3CA mutations in breast cancer patients from Central China Wu Haibo, Wang Wei, Du Jun, Li Hong, Wang Huogang, Huang Liangliang, Xiang Hang, Xie Jing, Liu Xiaoli, Li Heng, Lin Wenchu. Cancer Management and Research.2019;11. CrossRef
- Analysis of PIK3CA mutations in breast cancer subtypes Arsenic Ruza, Lehmann Annika, Budczies Jan, Koch Ines, Prinzler Judith, Kleine-Tebbe Anke, Schewe Christiane, Loibl Sibylle, Dietel Manfred, Denkert Carsten. Applied immunohistochemistry & molecular morphology: AIMM.2014;22(1). CrossRef
- Breast Cancer Survival Defined by the ER/PR/HER2 Subtypes and a Surrogate Classification according to Tumor Grade and Immunohistochemical Biomarkers Parise Carol A., Caggiano Vincent. Journal of Cancer Epidemiology.2014;2014. CrossRef
- Differences in Breast Cancer Survival by Molecular Subtypes in the United States Howlader Nadia, Cronin Kathleen A., Kurian Allison W., Andridge Rebecca. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology.2018;27(6). CrossRef
- Strategies for developing Ki67 as a useful biomarker in breast cancer Denkert Carsten, Budczies Jan, Minckwitz Gunter, Wienert Stephan, Loibl Sibylle, Klauschen Frederick. Breast (Edinburgh, Scotland).2015;24 Suppl 2. CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Biology , 2022
Author Details