Immunohistochemical Detection of BRAFV600E in Cutaneous Melanocytic Neoplasms in a Cohort of Egyptian Population
Download
Abstract
Objective: Melanocytic neoplasia shows different pathways that have been implicated in tumorigenesis. One of the key genes involved in the BRAFV600E gene, can be detected through DNA-based or protein-based tests including immunohistochemistry (IHC). To check its reproducibility, this study was conducted to evaluate BRAFV600E IHC by different observers in a range of melanocytic neoplasms.
Methods: The present study was performed to evaluate immunohistochemical staining (IHC) using the RM-08 to detect BRAF V600E in 50 formalin-fixed paraffin-embedded (FFPE) blocks of different melanocytic neoplasms after bleaching using Tris-HCL buffer. IHC was considered positive when more than 90% of tumour cells showed cytoplasmic staining. IHC assessment by a team of two pathologists and a third pathologist to check the interobserver variability. IHC scores were compared among nevi and melanoma.
Results: IHC evaluation revealed good agreement by different observers (ĸ-coefficient=0.691, p=0.00). BRAFV600E showed a statistically significant difference between nevi and melanoma.
Conclusions: These findings suggest that BRAFV600E IHC assessment shows good reproducibility among pathologists, however, more strict criteria should be adopted in interpretation, especially in bleached samples.
Introduction
Melanocytic neoplasms can be classified into benign nevi, melanocytomas and malignant melanomas on basis of biological behaviour. The incidence varies dramatically across different regions. Melanoma is the fifth most common malignancy in males in the USA [1]. Melanoma accounts for only 0.24% of malignancies in Egypt [2]. Pathogenesis depends on the interaction between environmental factors and host susceptibility. This has led to a paradigm shift in the classification model of melanocytic neoplasms which was issued in the 4th edition of the WHO classification of skin tumours. This model used the UV load, sun exposure (intermittent versus chronic), driver mutations and anatomic site [3, 4]. These models were based on Fairskinned individuals. However, the published data are few in the Middle East and North Africa (MENA) region, including the Egyptian population [2].
One of the key genes involved in nevi and melanoma formation is the BRAF gene. It is a human proto-oncogene located on the long arm of chromosome 7 (7q34) which encodes BRAF protein. The BRAF protein is a 766–amino acid-long protein formed of two regulatory domains and a kinase-encoding domain. BRAF protein is a member of the Raf kinase family of proteins which is a key regulator of the MAPK/ERK signalling pathway [3].
The BRAF protein constitutively activates MEK and ERK via phosphorylation leading to, uncontrolled stimulation of cell proliferation [4]. To date, more than 30 mutations of the BRAF gene associated with human cancers have been identified, the most common of which is the BRAFV600E mutation, in which hydrophilic glutamic acid (E) substitutes hydrophobic valine (V) at codon 600. Less commonly encountered mutations are grouped as BRAF non-V600E mutations [5].
Mutational status is identified via molecular testing using different techniques, including the FDA-approved Cobas test, sequencing, and real-time PCR. Immunohistochemistry using a monoclonal antibody VE1 clone has been proposed as a surrogate for RT–PCR molecular testing [5-7]. However, interpretation of immunohistochemical staining is evaluated by multiple methods with interobserver variability.
In this study, BRAF V600E was assessed using IHC in 50 formalin-fixed paraffin-embedded (FFPE) sections of different melanocytic neoplasms by three observers to identify BRAF V600E status and limit the interobserver variability. The results of IHC testing were compared between melanoma and nevi to evaluate the diagnostic value of BRAF V600E.
Materials and Methods
Patients
The current work included 50 retrospective excisional or incisional biopsies of 29 melanoma cases retrieved from the Pathology Laboratory archives, Faculty of Medicine, Alexandria University starting from January 2017 to January 2020. Ethical approval was granted by the Faculty of Medicine Ethics Committee (IRB no. 00007555, FWA NO. 00015712).
Immunohistochemical staining for BRAFV600E
Four-micrometre-thick sections were cut from paraffin blocks and placed on coated slides. Melanin bleaching for 29 moderately and heavily pigmented cases using 0.5% diluted hydrogen peroxide (H2O2) in Tris-HCl and PBS was performed as described by Chung et al [8] Antigen retrieval was performed using sodium citrate buffer in a microwave oven for 10 minutes. IHC staining was performed using Clone: RM-08, ThermoFisher Scientific, USA, dilution 1:300 in PBS according to the manufacturer’s protocol using a HrP kit (ThermoFisher Scientific) and diaminobenzidine (DAB) as a chromogen. positive and negative controls in the form of prostatic tissue and sections without primary antibody incubation, respectively.
Statistical Analysis
Statistical analysis was performed using SPSS version 20 (IBM) to evaluate interobserver variability and check the difference in BRAFV600E expression between nevi and melanoma.
Results
Immunohistochemistry for BRAF in Melanoma
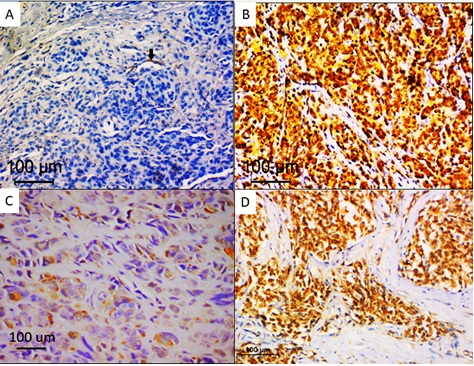
Out of 29 melanoma cases, melanin bleaching before immunohistochemical staining was performed in moderately and heavily pigmented cases. In 16 cases with prior bleaching, BRAF immunohistochemical staining was labelled as positive in 9 cases. In 13 cases, with absent or minimal pigmentation. eight cases revealed positive staining in the form of cytoplasmic staining in tumour cells with a total of 17 melanoma cases positive for BRAF immunostaining. The staining was almost homogeneous throughout tumour cells. Heterogeneous staining between different tumour regions was observed in the cases with prior bleaching, areas of necrosis, fixation artefacts or sometimes different tumour cell morphology. Ten cases were considered negative due to the complete absence of staining or patchy staining of scattered tumour cells and interspersed macrophages. Only two cases were considered ambiguous, one case in which prior bleaching was done revealed intense nuclear and cytoplasmic staining and staining of wide necrotic areas. Nuclear staining was observed in the overlying epidermis and cytoplasmic staining in endogenous blood vessels (positive internal control). Out of 21 nevi cases, only two cases showed positive cytoplasmic staining in tumour cells, one case was labelled histologically as pigmented epithelioid melancytoma and revealed cytoplasmic staining in more than 80% of tumour cells after melanin pigment bleaching. The second case was dermal nevus with verrucous overlying epidermis that showed minimal pigmentation (score +1), this case showed moderately intense cytoplasmic staining. Nineteen cases showed absent staining or patchy weak staining in less than 5% of nevus cells. Different IHC staining patterns are shown in Figure1.
Figure 1. BRAFV600E Immunohistochemical Staining in Melanoma. A, Negative staining in tumour cells with positive internal control (black arrow); B, Ambiguous staining in tumour cells; C, heterogeneous staining in tumour cells; D, diffuse homogenous staining in tumour cells. (immunoperoxidase X200).
Immunohistochemistry assessment by an independent observer
Using the same methodology, the third observer assessed the immunohistochemical staining in melanoma and nevi cases. Calculation of interobserver agreement between both teams using ĸ-agreement coefficient revealed good agreement) (ĸ-coefficient=0.691, p=0.00). Immunohistochemical staining scores assigned by different observers are summarized in Table 1.
| 3 rd observer scores | IHC by two observers | ||||
| Negative | Positive | Ambiguous | Total | ||
| Negative | Count | 25 (86.2) | 3 (15.8) | 1 (50.0) | 29 (58.0) |
| Positive | Count | 3 (10.3) | 16 (84.2) | 0 (0.0) | 19 (38.0) |
| Ambiguous | Count | 1 (3.4) | 0 | 1 (50.0) | 24 |
| Total | Count | 29 | 19 | 2 | 50 |
Difference between BRAFV600E expression between nevi and melanoma
On Comparison between BRAFV600E IHC results in nevi and melanoma cases using the chi-square test, there was a statistically significant difference in PCR results between melanoma and nevi as 9.5% of nevi and 58.6% of melanoma cases were positive (i.e. expressing mutant BRAFV600E). (P (MC)=0.00) (Table 2).
| Immunohistochemistry results | Nevi | Melanoma | Total | |
| Negative | Count | 19 (90.5) | 10 (34.5) | 29 (58.0) |
| Positive | Count | 2 (9.5) | 17 (58.6) | 19 (38.0) |
| Ambiguous | Count | 0 | 2 (6.9) | 2 (4.0) |
| Total | Count | 21 | 29 | 50 |
The Immunohistochemistry staining results were validated using BRAFV600E CAST-PCR (data not shown) and revealed a sensitivity of 66.7%, specificity of 73.3%, a positive predictive value of 72.22%, a negative predictive value of 84.6%, Overall agreement of 71.43%.
Discussion
BRAFV600E mutational analysis assessment is crucial to identify patients’ eligibility for BRAF kinase inhibitors. BRAFV600E status is assessed using molecular testing by sequencing-based techniques. Recently the use of monoclonal antibodies specific to the mutation has been proposed as a surrogate marker [7, 9].
In the present work, Immunohistochemistry was performed and revealed lower sensitivity and specificity in BRAFV600E compared to the published series. This could be attributed to variation in preanalytic, analytic and post-analytical parameters. Pre-analytic includes variable specimen size, cold ischaemia, and fixation process [10].
In the analytic phase, bleaching before IHC and the use of different antibody clones (RM-8) may affect the process. Zhang et al [11] reported false negative staining when prior bleaching was employed before VE1 [7, 12, 13]. Monoclonal antibodies apart from the VE1 clone have been reported to have lower specificity and sensitivity in BRAFV600E mutation detection [14].
In the post-analytic phase, training on IHC interpretation is crucial to avoid interobserver variability [6, 13]. Since 2011, multiple methods have been used to interpret the staining. The most commonly used was proposed by Capper et al [7, 13] and was used in the current work. Although, the method showed good reproducibility. Interpretation of heavily pigmented melanomas remained problematic, and a more strict method was proposed by Fisher etal [6]. Recently, a meta-analysis showed that intratumoral heterogeneity in BRAFV600E IHC might explain its lower sensitivity in BRAFV600E mutation detection [15]. Yancovitz etal [16] reported that intratumoral heterogeneity in BRAF expression caused a marked discrepancy between BRAFV600E molecular testing methods.
Nevertheless, the use of PCR-based tests was reported to be a more rapid, sensitive, specific and cost-effective method for detecting BRAF mutations [17-20].
In the present study, BRAF IHC expression showed a statistically significant difference between nevi and melanoma. BRAF V600E expression was found to be associated with tumour progression and a predictive marker of BRAF inhibitors [21].
In conclusions, the results obtained in this study indicate that the IHC method can be used as a screening tool for BRAFV600E; however, cannot be used as a surrogate marker for based tests including CAST-PCR and sequencing. There is no consensus on BRAF IHC staining interpretation criteria among different study groups, which in turn questions the methodology that should be adopted for staining interpretation.
Acknowledgements
General
The authors would like to thank Asmaa Abdelhameed for consultation on statistical analysis.
Funding Statement
The research is funded by the research team.
Approval
The research is approved by the faculty of medicine, at Alexandria university.
Conflict of Interest
None declared.
Ethical Declaration
Clinical data were collected and evaluated according to the approval of the Faculty of Medicine Ethics Committee, Alexandria University (IRB no. 00007555, FWA NO. 00015712).). The study was conducted by the international standards of good clinical practice and with the 1964 Helsinki declaration and its later amendments.
Authors Contribution
Nada M. Yakout, Dina M. Abdallah, Doaa Abdelmonsif and Omayma El-Sakka participated in the study design and coordination. Nada M Yakout, Omayma El-Sakka and Dina M. Abdallah performed the immunohistochemical analysis. Doaa Abedimonsif performed and interpreted CAST-PRC for data validation. Hassan Mahmoud Kholosy collected and provided all clinical data. Nada M Yakout and Omayma El-Sakka wrote the main manuscript text. Nada M. Yakout and Dina M. Abdallah Omayma El-Sakka prepared Figure 1, The authors have read and approved the final manuscript.
Data Availability
Raw data sharing does not apply to Immunohistochemistry as no datasets were generated.
References
- Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries Sung H, Ferlay J, Siegel RL , Laversanne M, Soerjomataram I, Jemal A, Bray F. CA: a cancer journal for clinicians.2021;71(3). CrossRef
- Skin cancer in Egypt: a word in your ear Hussein MR . Cancer Biology & Therapy.2005;4(5). CrossRef
- MAP kinases Chen Z., Gibson T. B., Robinson F., Silvestro L., Pearson G., Xu B., Wright A., Vanderbilt C., Cobb M. H.. Chemical Reviews.2001;101(8). CrossRef
- Structure of the BRAF-MEK complex reveals a kinase activity independent role for BRAF in MAPK signaling Haling JR , Sudhamsu J, Yen I, Sideris S, Sandoval W, Phung W, Bravo BJ , Giannetti AM , Peck A, Masselot A, Morales T, Smith D, Brandhuber BJ , Hymowitz SG , Malek S. Cancer Cell.2014;26(3). CrossRef
- Molecular testing for BRAF mutations to inform melanoma treatment decisions: a move toward precision medicine Cheng L, Lopez-Beltran A, Massari F, MacLennan GT , Montironi R. Modern Pathology: An Official Journal of the United States and Canadian Academy of Pathology, Inc.2018;31(1). CrossRef
- Accurate detection of BRAF p.V600E mutations in challenging melanoma specimens requires stringent immunohistochemistry scoring criteria or sensitive molecular assays Fisher KE , Cohen C, Siddiqui MT , Palma JF , Lipford EH , Longshore JW . Human Pathology.2014;45(11). CrossRef
- Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody Capper D, Preusser M, Habel A, Sahm F, Ackermann U, Schindler G, Pusch S, Mechtersheimer G, Zentgraf H, Deimling A. Acta Neuropathologica.2011;122(1). CrossRef
- A melanin-bleaching methodology for molecular and histopathological analysis of formalin-fixed paraffin-embedded tissue Chung J, Choi J, Sears JD , Ylaya K, Perry C, Choi CH , Hong S, Cho H, Brown KM , Hewitt SM . Laboratory Investigation; a Journal of Technical Methods and Pathology.2016;96(10). CrossRef
- Utility of BRAF V600E Immunohistochemistry Expression Pattern as a Surrogate of BRAF Mutation Status in 154 Patients with Advanced Melanoma Tetzlaff MT , Pattanaprichakul P, Wargo J, Fox PS , Patel KP , Estrella JS , Broaddus RR , Williams MD , Davies MA , Routbort MJ , Lazar AJ , Woodman SE , Hwu W , Gershenwald JE , Prieto VG , Torres-Cabala CA , Curry JL . Human Pathology.2015;46(8). CrossRef
- Immunohistochemistry with the anti-BRAF V600E (VE1) antibody: impact of pre-analytical conditions and concordance with DNA sequencing in colorectal and papillary thyroid carcinoma Dvorak K, Aggeler B, Palting J, McKelvie P, Ruszkiewicz A, Waring P. Pathology.2014;46(6). CrossRef
- A validation study for the use VE1 immunohistochemical staining in screening for BRAF mutation in cutaneous malignant melanoma Zhang W, Song G, Han X, Li X, Luo D. Biomedical Research.2017;28(11):4886-4890.
- Immunohistochemistry as a reliable method for detection of BRAF-V600E mutation in melanoma: a systematic review and meta-analysis of current published literature Anwar MAF , Murad F, Dawson E, Abd Elmageed ZY , Tsumagari K, Kandil E. The Journal of Surgical Research.2016;203(2). CrossRef
- Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma Long GV , Wilmott JS , Capper D, Preusser M, Zhang YE , Thompson JF , Kefford RF , Deimling A, Scolyer RA . The American Journal of Surgical Pathology.2013;37(1). CrossRef
- Comparison of 2 monoclonal antibodies for immunohistochemical detection of BRAF V600E mutation in malignant melanoma, pulmonary carcinoma, gastrointestinal carcinoma, thyroid carcinoma, and gliomas Routhier CA , Mochel MC , Lynch K, Dias-Santagata D, Louis DN , Hoang MP . Human Pathology.2013;44(11). CrossRef
- BRAF Heterogeneity in Melanoma Ito T, Tanaka Y, Murata M, Kaku-Ito Y, Furue K, Furue M. Current Treatment Options in Oncology.2021;22(3). CrossRef
- Intra- and inter-tumor heterogeneity of BRAF(V600E))mutations in primary and metastatic melanoma Yancovitz M, Litterman A, Yoon J, Ng E, Shapiro RL , Berman RS , Pavlick AC , Darvishian F, Christos P, Mazumdar M, Osman I, Polsky D. PloS One.2012;7(1). CrossRef
- BRAF V600 mutation detection in melanoma: a comparison of two laboratory testing methods O'Brien O, Lyons T, Murphy S, Feeley L, Power D, Heffron CCBB . Journal of Clinical Pathology.2017;70(11). CrossRef
- Sensitive allele-specific real-time PCR test for mutations in BRAF codon V600 in skin melanoma Pisareva E, Gutkina N, Kovalenko S, Kuehnapfel S, Hartmann A, Heinzerling L, Schneider-Stock R, Lyubchenko L, Shamanin VA . Melanoma Research.2014;24(4). CrossRef
- Prospective evaluation of two screening methods for molecular testing of metastatic melanoma: Diagnostic performance of BRAF V600E immunohistochemistry and of a NRAS-BRAF fully automated real-time PCR-based assay Vallée A, Denis-Musquer M, Herbreteau G, Théoleyre S, Bossard C, Denis MG . PLoS ONE.2019;14(8). CrossRef
- A multisite blinded study for the detection of BRAF mutations in formalin-fixed, paraffin-embedded malignant melanoma Richter A, Grieu F, Carrello A, Amanuel B, Namdarian K, Rynska A, Lucas A, Michael V, Bell A, Fox SB , Hewitt CA , Do H, McArthur GA , Wong SQ , Dobrovic A, Iacopetta B. Scientific Reports.2013;3. CrossRef
- A combination of p300 and Braf expression in the diagnosis and prognosis of melanoma Bhandaru M, Ardekani GS , Zhang G, Martinka M, McElwee KJ , Li G, Rotte A. BMC cancer.2014;14. CrossRef
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Copyright
© Asian Pacific Journal of Cancer Care , 2022
Author Details
How to Cite
- Abstract viewed - 0 times
- PDF (FULL TEXT) downloaded - 0 times
- XML downloaded - 0 times